
We will begin with DNA, which is the hereditary information in every cell, that is copied and passed on from generation to generation.
DNA was discovered in 1869 by Friedrich Miescher, and was identified as the genetic material in experiments in the 1940s led by Oswald Avery, Colin MacLeod, and Maclyn McCarty.
In 1953, the international Science journal Nature published 3 papers on the structure of DNA. The authors included James Watson and Francis Crick, Rosalind Franklin and Erwin Chargaff. Arguably, the Watson and Crick paper has had more scientific impact per word than any other research article ever published. Today, instruction about the double helical structure of DNA is foundational to biology classes in high schools and colleges. It launched the study of genetics forward by providing information that linked structure at the atomic and molecular level to heredity and the basis of inherited disease.
The double helix structure presented in the Watson and Crick paper and justified by the X-ray data from Rosalind Franklin, includes a 2-strand structure which has at its core, fused ring structures called nitrogenous bases joined by hydrogen bonds to form base pairs. The base adenine always pairs with thymine, and the base guanine invariably pairs with cytosine. Two hydrogen bonds form between adenine and thymine, and three hydrogen bonds hold together guanine and cytosine (Figure 2.127).
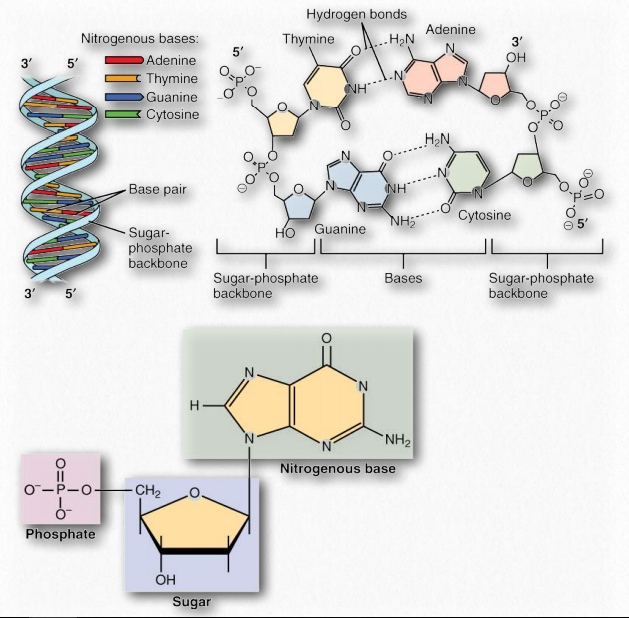
Figure 2.127 – A DNA duplex with base pairs, a closeup of base pairing, and a closeup of a nucleotide Wikipedia
The complementary structure of these bases revealed to Watson and Crick how DNA might be (and in fact, is) replicated. Further, it indicates how information is DNA is transmitted to RNA for the synthesis of proteins. Both of these important biological processes rely on the ability of either strand of the double helix being ‘readable’ in isolation from the other. Separation of the strand exposes hydrogen bonding sites where a complementary strand could be built up, purely by knowing the basis of the pairing: A pairs with T, G with C, etc..
Molecular machinery does essentially this, as it uses a single strand as a template for reproducing DNA molecules with the proper base sequences. Separate molecular machinery also transcribes this information to mRNA, where the code can then be translated into an amino acid sequence. Strings of amino acids fold into proteins, and thus the stored information finds expression as functioning proteins.
Structure
A single DNA strand (half of a double helix) can be described as a polymer of nucleoside monophosphates held together by phosphodiester bonds. It is a long and narrow molecule. Two such paired strands make up the DNA molecule, which is then twisted into a helix. In the most common B form, the DNA helix twists with a repeat of 10.5 base pairs per full helical turn, with sugars and phosphate forming the covalent phosphodiester “backbone” of the molecule and the adenine, guanine, cytosine, and thymine bases oriented in the middle where they form the now familiar base pairs that look like the rungs of a ladder.
Building blocks
The term nucleotide refers to the building blocks of both DNA (deoxyribonucleoside triphosphates, dNTPs) and RNA (ribonucleoside triphosphates, NTPs). In order to discuss this important group of molecules, it is necessary to define some terms.
Nucleotides contain three primary structural components. These are a nitrogenous base, a 5-carbon pentose sugar, and at least one phosphate. The nitrogenous bases found in nucleic acids include adenine and guanine (called purines) and cytosine, uracil, or thymine (called pyrimidines).
There are two sugars found in nucleotides – deoxyribose and ribose (Figure 2.128). Deoxyribose differs from ribose at carbon 2, with ribose having an OH group, where deoxyribose has H.
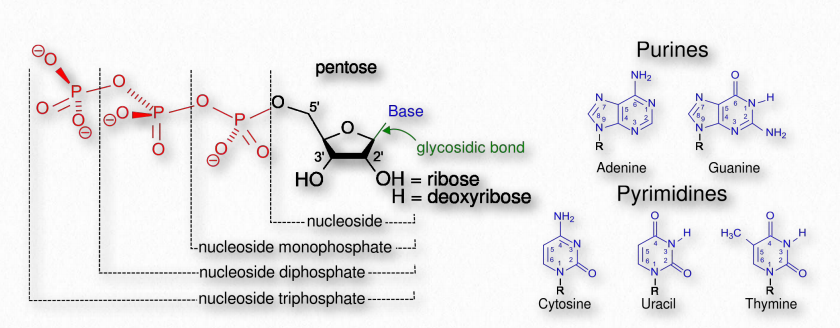
Figure 2.128 – Nucleotides, nucleosides, and bases
Nucleotides containing deoxyribose are called deoxyribonucleotides and are the forms found in DNA.
Nucleotides containing ribose are called ribonucleotides and are found in RNA.
Both DNA and RNA contain nucleotides with adenine, guanine, and cytosine, but with very minor exceptions, RNA contains uracil nucleotides, whereas DNA contains thymine nucleotides. When a base is attached to a sugar, the product gains a new name.
- uracil-containing = uridine (attached to ribose) / deoxyuridine (attached to deoxyribose)
- thymine-containing = ribothymidine (attached to ribose) / thymidine (attached to deoxyribose)
- cytosine-containing = cytidine (attached to ribose – Figure 2.129) / deoxycytidine (attached to deoxyribose)
- guanine-containing = guanosine (attached to ribose) / deoxyguanosine (attached to deoxyribose)
- adenine-containing = adenosine (attached to ribose) / deoxyadenosine (attached to deoxyribose)
The addition of one or more phosphates to a nucleoside makes it a nucleotide. Nucleotides are often referred to as nucleoside phosphates, for this reason. The number of phosphates in the nucleotide is indicated by the appropriate prefixes (mono, di or tri).
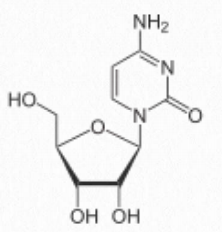
Figure 2.129 Cytidine
Addition of second and third phosphates to a nucleoside monophosphate requires an input of energy, due to the repulsion of negatively charged phosphates. This chemical energy can be released in the reverse reaction. This conversion of triphosphate nucleotides (such as ATP) to diphosphates is a reaction that is employed for energy storage and utilization by cells.
Note: Ribonucleotides as Energy Sources
Though ATP is the most common and best known cellular energy source, each of the four ribonucleotides plays important roles in providing energy. GTP, for example, is the energy source for protein synthesis (translation) as well as for a handful of metabolic reactions.
Hydrogen bonds
Hydrogen bonds between the base pairs hold a nucleic acid duplex together, with two hydrogen bonds per A-T pair (or per A-U pair in RNA) and three hydrogen bonds per G-C pair. The B-form of DNA has a prominent major groove and a minor groove tracing the path of the helix (Figure 2.132). Proteins, such as transcription factors bind in these grooves and access the hydrogen bonds of the base pairs to “read” the sequence therein.
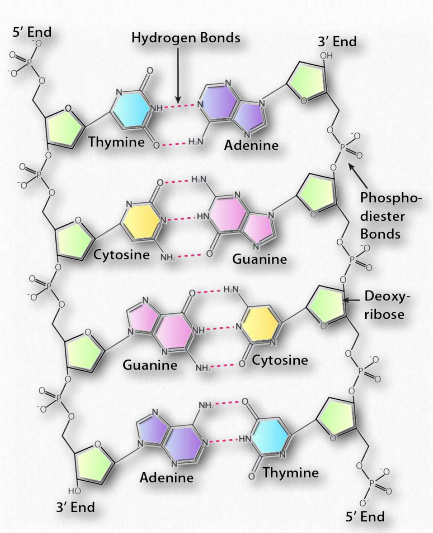
Figure 2.131 – Anti-parallel orientation of a DNA duplex, phosphodiester backbone, and base pairing Image by Aleia Kim
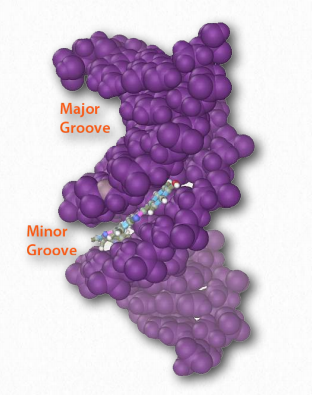
Figure 2.132 – Major and minor grooves of DNA. The minor groove has been bound by a dye Wikipedia
Movie 2.5 – B-form DNA duplex rotating in space Wikipedia
RNA
The structure of RNA (Figure 2.137) is very similar to that of a single strand of DNA. Built of ribonucleotides, joined together by the same sort of phosphodiester bonds as in DNA, RNA uses uracil in place of thymine. The building of messenger RNAs by copying a DNA template is a crucial step in the transfer of the information in DNA to a form that directs the synthesis of protein. Additionally, ribosomal and transfer RNAs serve important roles in “reading” the information in the mRNA codons and in polypeptide synthesis. RNAs are also known to play important roles in the regulation of gene expression.
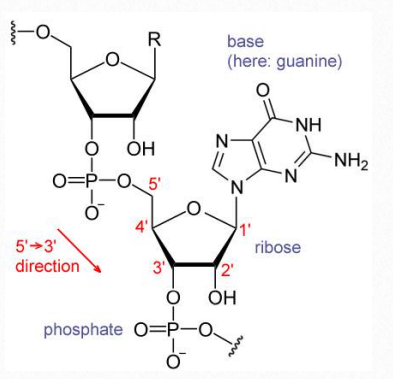
Figure 2.137 A section of an RNA molecule Wikipedia
RNA world
The discovery, in 1990, that RNAs could play a role in catalysis, a function once thought to be solely the domain of proteins, was followed by the discovery of many more so-called ribozymes- RNAs that functioned as enzymes. This suggested the answer to a long-standing chicken or egg puzzle – if DNA encodes proteins, but the replication of DNA requires proteins, how did a replicating system come into being? This problem could be solved if the first replicator was RNA, a molecule that can both encode information and carry out catalysis. This idea, called the “RNA world” hypothesis, suggests that DNA as genetic material and proteins as catalysts arose later, and eventually prevailed because of the advantages they offer. The lack of a 2’OH on deoxyribose makes DNA more stable than RNA. The double-stranded structure of DNA also provides an elegant way to easily replicate it. RNA catalysts, however, remain, as remnants of that early world. In fact, the formation of peptide bonds, essential for the synthesis of proteins, is catalyzed by RNA.
Secondary structure
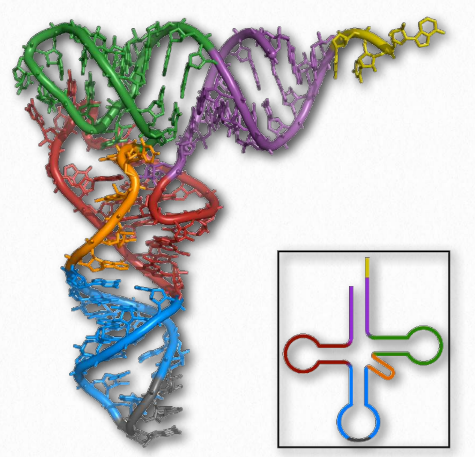
Figure 2.138 – tRNA Images – 3D projection (left) and 2D projection (inset) Wikipedia
With respect to structure, RNAs are more varied than their DNA cousin. Created by copying regions of DNA, cellular RNAs are synthesized as single strands, but they often have self-complementary regions leading to “foldbacks” containing duplex regions. These are most easily visualized in the ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) (Figure 2.138), though other RNAs, including messenger RNAs (mRNAs), small nuclear RNAs (snRNAs), microRNAs (Figure 2.139), and small interfering RNAs (siRNAs) may each have double helical regions as well.

Figure 2.139 – MicroRNA stem loops. Wikipedia
Base pairing
Base pairing in RNA is slightly different than DNA. This is due to the presence of the base uracil in RNA in place of thymine in DNA. Like thymine, uracil forms base pairs with adenine, but unlike thymine, uracil can, to a limited extent, also base pair with guanine, giving rise to many more possibilities for pairing within a single strand of RNA.
These additional base pairing possibilities mean that RNA has many ways it can fold upon itself that single-stranded DNA cannot. Folding, of course, is critical for protein function, and we now know that, like proteins, some RNAs in their folded form can catalyze reactions just like enzymes. Such RNAs are referred to as ribozymes. It is for this reason scientists think that RNA was the first genetic material, because it could not only carry information, but also catalyze reactions. Such a scheme might allow certain RNAs to make copies of themselves, which would, in turn, make more copies of themselves, providing a positive selection.
Stability
RNA is less chemically stable than DNA. The presence of the 2’ hydroxyl on ribose makes RNA much more prone to hydrolysis than DNA, which has a hydrogen instead of a hydroxyl. Further, RNA has uracil instead of thymine. It turns out that cytosine is the least chemically stable base in nucleic acids. It can spontaneously deaminate and in turn is converted to a uracil. This reaction occurs in both DNA and RNA, but since DNA normally has thymine instead of uracil, the presence of uracil in DNA indicates that deamination of cytosine has occurred and that the uracil needs to be replaced with a cytosine. Such an event occurring in RNA would be essentially undetectable, since uracil is a normal component of RNA. Mutations in RNA have much fewer consequences than mutations in DNA because they are not passed between cells in division.
Catalysis
RNA structure, like protein structure, has importance, in some cases, for catalytic function. Like random coils in proteins that give rise to tertiary structure, single-stranded regions of RNA that link duplex regions give these molecules a tertiary structure, as well. Catalytic RNAs, called ribozymes, catalyze important cellular reactions, including the formation of peptide bonds in ribosomes (Figure 2.114). DNA, which is usually present in cells in strictly duplex forms (no tertiary structure, per se), is not known to be involved in catalysis.
RNA structures are important for reasons other than catalysis. The 3D arrangement of tRNAs is necessary for enzymes that attach amino acids to them to do so properly. Further, small RNAs called siRNAs found in the nucleus of cells appear to play roles in both gene regulation and in cellular defenses against viruses. The key to the mechanisms of these actions is the formation of short foldback RNA structures that are recognized by cellular proteins and then chopped into smaller units. One strand is copied and used to base pair with specific mRNAs to prevent the synthesis of proteins from them.
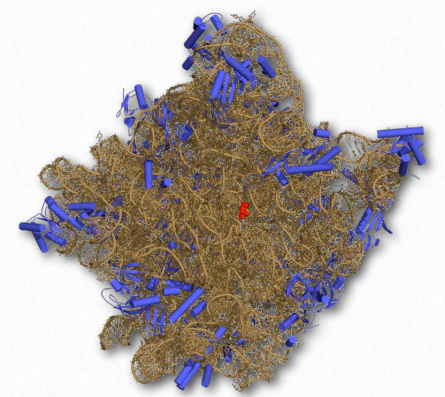
Figure 2.140 – Structure of the 50S ribosomal subunit. rRNA shown in brown. Active site in red Wikipedia
Ames test
The Ames test (Figure 2.147) is an analytical method that allows one to determine whether a compound causes mutations in DNA (is mutagenic) or not. The test is named for Dr. Bruce Ames, a UC Berkeley emeritus professor who was instrumental in creating it. In the procedure, a single base pair of a selectable marker of an organism is mutated in a plasmid to render it nonfunctional. In the example, a strain of Salmonella is created that lacks the ability to grow in the absence of histidine. Without histidine, the organism will not grow, but if that one base in the plasmid’s histidine gene gets changed back to its original base, a functional gene will be made and the organism will be able to grow without histidine.
A culture of the bacterium lacking the functional gene is grown with the supply of histidine it requires. It is split into two vials. To one of the vials, a compound that one wants to test the mutagenicity of is added. To the other vial, nothing is added. The bacteria in each vial are spread onto plates lacking histidine. In the absence of mutation, no bacteria will grow. The more colonies of bacteria that grow, the more mutation happened. Note that even the vial without the possible mutagenic compound will have a few colonies grow, as a result of mutations unlinked to the potential mutagen.
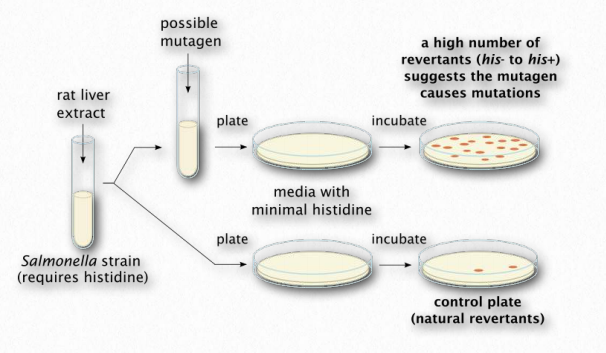
Figure 2.147 – The Ames test Wikipedia
Mutation happens in all cells at a low level. If the plate with the cells from the vial with the compound has more colonies than the cells from the control vial (no compound), then that would be evidence that the compound causes more mutations than would normally occur and it is therefore a mutagen. On the other hand, if there was no significant difference in the number of colonies on each plate, then that would suggest it is not mutagenic. The test is not perfect – it identifies about 90% of known mutagens – but its simplicity and inexpensive design make it an excellent choice for an initial screen of a compound.

