
In eukaryotic cells, the vast majority of ATP synthesis occurs in the mitochondria in a process called oxidative phosphorylation. Even plants, which generate ATP by photophosphorylation in chloroplasts, contain mitochondria for the synthesis of ATP through oxidative phosphorylation.
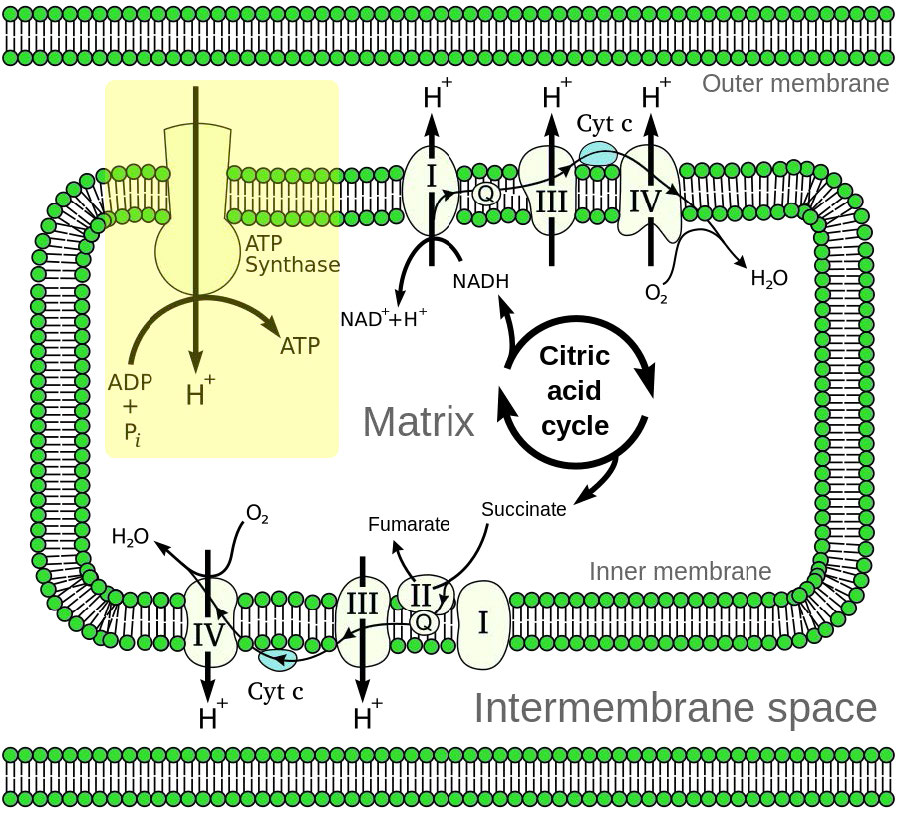
Oxidative phosphorylation is linked to a process known as electron transport (Figure 5.14). The electron transport system (also called the Electron Transport Chain, or ETC), located in the inner mitochondrial membrane, transfers electrons donated by the reduced molecules NADH and FADH2 through a series of electrons acceptors, to oxygen. As we shall see, movement of electrons through system essentially sets up a gradient across a membrane that is then used to make ATP.
The NADH and FADH2 that acts as the fuel for this process comes from other parts of metabolism for energy. The end products are ATP and water.
Chemiosmotic model
Dr. Peter Mitchell introduced a radical proposal in 1961 to explain the mechanism by which mitochondria make ATP. It is known as the chemiosmotic hypothesis and has been shown over the years to be correct.
Mitchell proposed that synthesis of ATP in mitochondria depends on an electrochemical gradient, across the mitochondrial inner membrane, that arises ultimately from the energy of reduced electron carriers, NADH and FADH2.
There are three general processes that work together to accomplish this feat:
- Electrons are passed through the ETC: A pH gradient (also described as a proton gradient or hydrogen ion gradient) is created when electrons from NADH and FADH2 are passed through the electron transport chain (ETC) located in the inner mitochondrial membrane.
- Protons are pumped across a membrane, setting up a gradient: Movement of electrons through the series of of electron carriers is coupled to the pumping of protons out of the mitochondrial matrix across the inner mitochondrial membrane.
- ATP Synthase catalyzes phosphorylation of ADP: protons re-enter the mitochondrial matrix via the transmembrane ATP synthase protein complex, which combines ADP with inorganic phosphate to make ATP.
Tight coupling
Under normal metabolic conditions, tight coupling is said to exist between electron transport and the synthesis of ATP (called oxidative phosphorylation). Chemicals which permeabilize the inner mitochondrial membrane disallow the gradient to form, causing uncoupling of the ETC to ATP synthesis. That is, they allow the protons to leak back into the mitochondrial matrix, rather than through the ATP synthase, so that the movement of electrons through the ETS can no longer support the synthesis of ATP.
Such uncoupling occurs in situations such as:
- Brown fat in animals uses uncoupling to generate heat but not ATP when heat generation is a priority, such as during hibernation or in infants unable to maintain their temperature alone.
- Some plants use uncoupling to heat their tissues and volatilize odor chemicals that attract pollinators.
- Some poisons, including insecticides, cause uncoupling and starve an organism from the production of ATP, leading to death.
Power plants
Mitochondria are called the power plants of the cell because most of a cell’s ATP is produced there via oxidative phosphorylation. The mechanism by which ATP is made in oxidative phosphorylation is one of the most interesting in all of biology.
Considerations
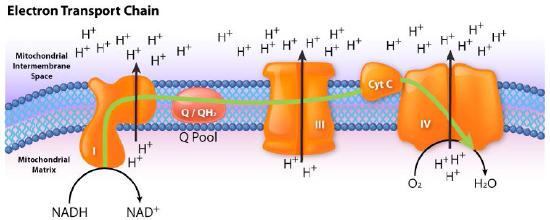
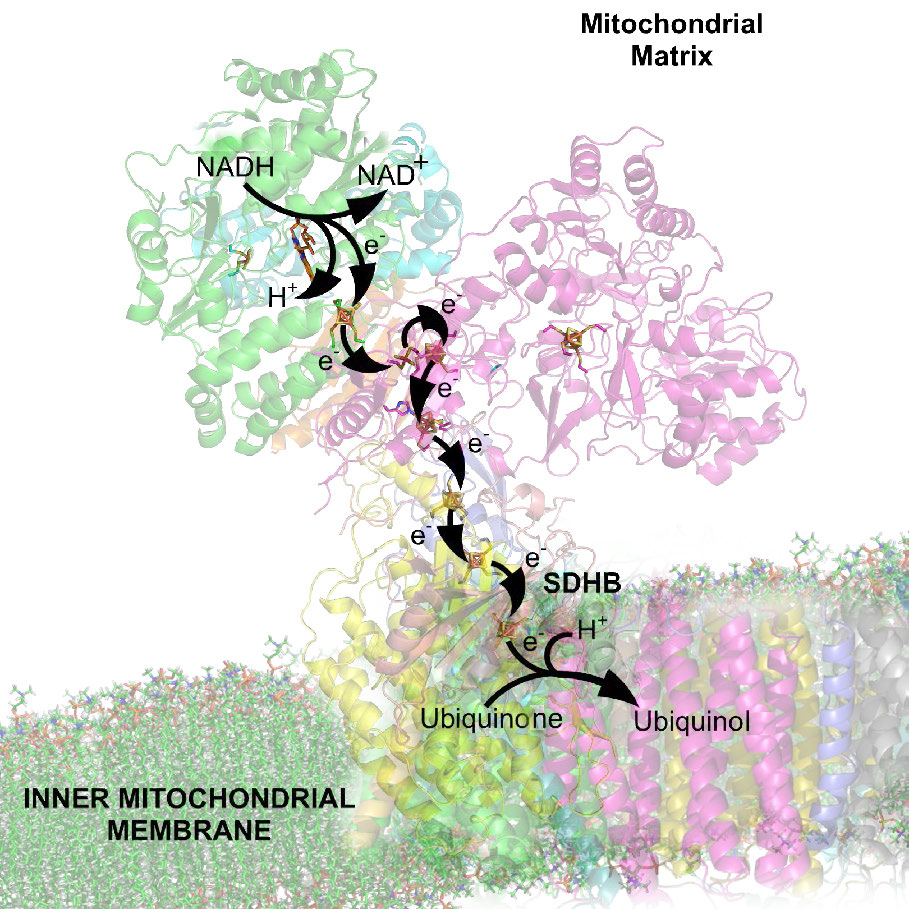
The process has three primary considerations. The first is electrical – electrons from reduced electron carriers, such as NADH and FADH2, enter the electron transport system via Complex I and II, respectively. As seen in Figure 5.16 and Figure 5.17, electrons move from one complex to the next, not unlike the way they move through an electrical circuit. Such movement occurs a a result of a set of reduction-oxidation (redox) reactions with electrons moving from a more negative reduction potential to a more positive one.
One can think of this occurring as a process where carriers “take” electrons away from complexes with lower reduction potential, much the way a bully takes lunch money from a smaller child. In this scheme, the biggest “bully” is oxygen in Complex IV. Electrons gained by a carrier cause it to be reduced, whereas the carrier giving up the electrons is oxidized.
Entry of electrons to system
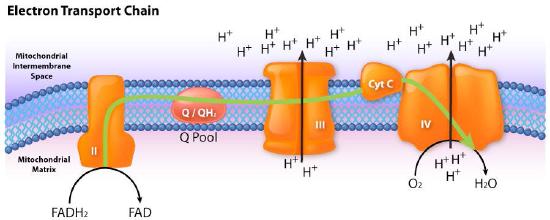
Movement of electrons through the chain begins either by 1) transfer from NADH to Complex I (Figure 5.16) or 2) movement of electrons through a covalently bound FADH2 (Figure 5.17) in the membrane-bound succinate dehydrogenase (Complex II). (An alternate entry point for electrons from FADH2 is the Electron Transferring Flavoprotein via the electron-transferring-flavoprotein dehydrogenase, not shown).
Traffic cop
Both Complex I and II pass electrons to the inner membrane’s coenzyme Q (CoQ – Figures 5.18 & 5.19). In each case, coenzyme Q accepts electrons in pairs and passes them off to Complex III (CoQH2-cytochrome c reductase) singly. Coenzyme Q thus acts as a traffic cop, regulating the flow of electrons through the ETS.
Docking station
Complex III is a docking station or interchange for the incoming electron carrier (coenzyme Q) and the outgoing carrier (cytochrome c). Movement of electrons from Coenzyme Q to Complex III and then to cytochrome C occurs as a result of what is referred to as the Q-cycle (see below).
Complex III acts to ferry electrons from CoQ to cytochrome c. Cytochrome c takes one electron from Complex III and passes it to Complex IV (cytochrome oxidase). Complex IV is the final protein recipient of the electrons. It passes them to molecular oxygen (O2) to make two molecules of water. Making two water molecules requires four electrons, so Complex IV must accept, handle, and pass to molecular oxygen four separate electrons, causing the oxidation state of oxygen to be sequentially changed with addition of each electron.
Proton pumping
As electrons pass through complexes I, III, and IV, there is a release of a small amount of energy at each step, which is used to pump protons from the mitochondrial matrix (inside of mitochondrion) and deposit them in the intermembrane space (between the inner and outer membranes of the mitochondrion). The effect of this redistribution is to increase the electrical and chemical potential across the membrane.
Potential energy
As discussed earlier, electrochemical gradients have potential energy. Students may think of the process as “charging the battery.” Just like a charged battery, the potential arising from the proton differential across the membrane can be used to do things. In the mitochondrion, what the proton gradient does is facilitate the production of ATP from ADP and Pi. This process is known as oxidative phosphorylation, because the phosphorylation of ADP to ATP is dependent on the chemical oxidations occurring in the mitochondria.
ATP synthase
The protein complex harvesting energy from the proton gradient and using it to make ATP from ADP is an enzyme that has several names – Complex V, PTAS (Proton Translocating ATP Synthase), and ATP synthase (Figure 5.29). Central to its function is the movement of protons through it (from the intermembrane space back into the matrix). Protons will only provide energy to make ATP if their concentration is greater in the intermembrane space than in the matrix and if ADP is available.
In summary, the electron transport system charges the battery for oxidative phosphorylation by pumping protons out of the mitochondrion. The intact inner membrane of the mitochondrion keeps the protons out, except for those that re-enter through ATP Synthase. The ATP Synthase allows protons to re-enter the mitochondrial matrix and harvests their energy to make ATP.
ATP synthase mechanism
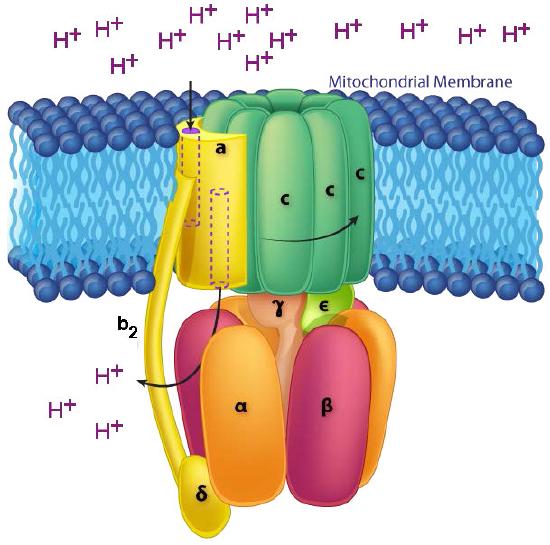
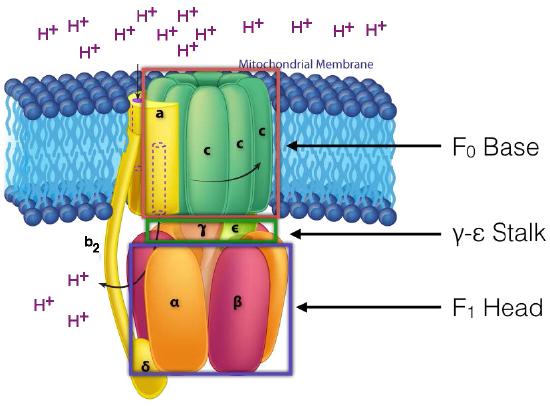
The ATP Synthase itself is an amazing nanomachine that makes ATP using a gradient of protons flowing through it from the intermembrane space back into the matrix. It is not easy to depict in a single image what the synthase does. Figure 5.31 and Figure 5.32 illustrate the multi-subunit nature of this membrane protein, which acts like a turbine at a hydroelectric dam. The movement of protons through the ATP Synthase c-ring causes it and the γ-ε stalk attached to it to turn. It is this action that is necessary for making ATP.
In ATP Synthase, the spinning components, or rotor, are the membrane portion (c ring) of the F0 base and the γ-ε stalk, which is connected to it. The γ-ε stalk projects into the F1 head of the mushroom structure. The F1 head contains the catalytic ability to make ATP. The F1 head is hexameric in structure with paired α and β proteins arranged in a trimer of dimers. ATP synthesis occurs within the β subunits.
Rotation of γ unit
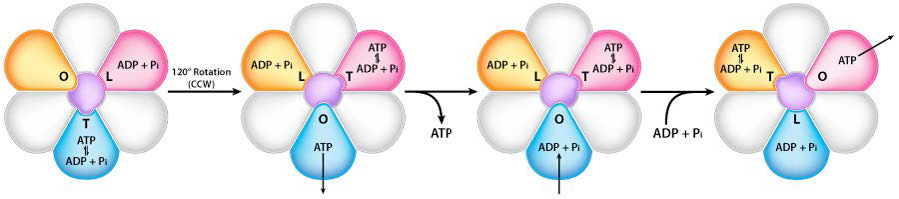
Turning of the γ shaft (caused by proton flow) inside the α-β trimer of the F1 head causes each set of β proteins to change structure slightly into three different forms called Loose, Tight, and Open (L,T,O – Figure 5.31). Each of these forms has a function.
The Loose form binds ADP + Pi. The Tight form “squeezes” them together to form the ATP. The Open form releases the ATP into the mitochondrial matrix. Thus, as a result of the proton flow through the ATP synthase, from the intermembrane space into the matrix, ATP is made from ADP and Pi.
Energy efficiency
Cells are not 100% efficient in energy use. Nothing we know is. Consequently, cells do not get as much energy out of catabolic processes as they put into anabolic processes. A good example is the synthesis and breakdown of glucose, something liver cells are frequently doing. The complete conversion of glucose to pyruvate in glycolysis (catabolism) yields two pyruvates plus 2 NADH plus 2 ATPs. Conversely, the complete conversion of two pyruvates into glucose by gluconeogenesis (anabolism) requires 4 ATPs, 2 NADH, and 2 GTPs. Since the energy of GTP is essentially equal to that of ATP, gluconeogenesis requires a net of 4 ATPs more than glycolysis yields. This difference must be made up in order for the organism to meet its energy needs. It is for this reason that we eat. In addition, the inefficiency of our capture of energy in reactions results in the production of heat and helps to keep us warm.
Metabolic controls of energy
It is also noteworthy that cells do not usually have both catabolic and anabolic processes for the same molecules occurring simultaneously inside of them (for example, breakdown of glucose and synthesis of glucose) because the cell would see no net production of anything but heat and a loss of ATPs with each turn of the cycle. Such cycles are called futile cycles and cells have controls in place to limit the extent to which they occur. Since futile cycles can, in fact, yield heat, they are used as sources of heat in some types of tissue. Brown adipose tissue of mammals uses this strategy, as described earlier. See also HERE for more on heat generation with a futile cycle.
Reactive oxygen species
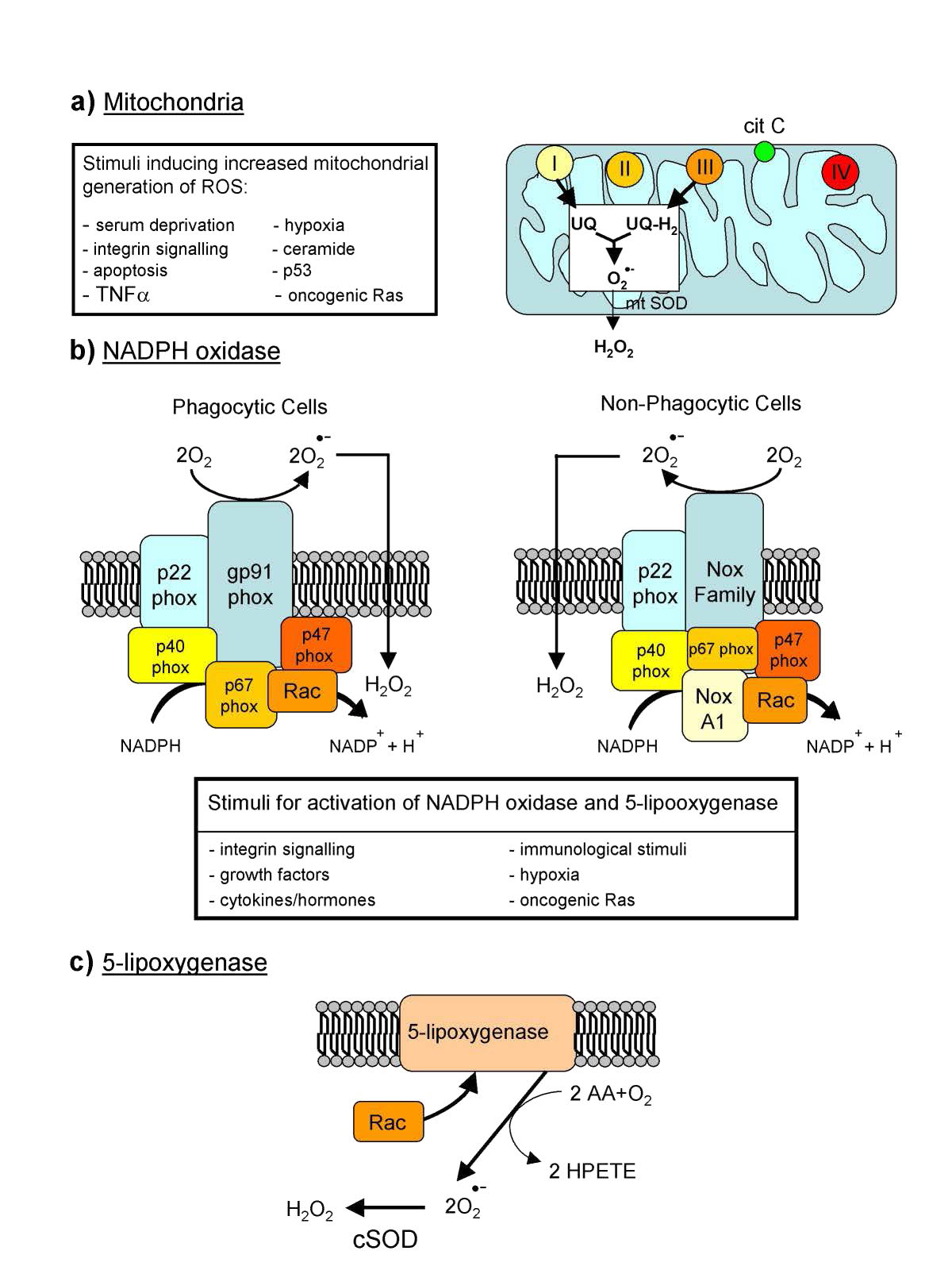
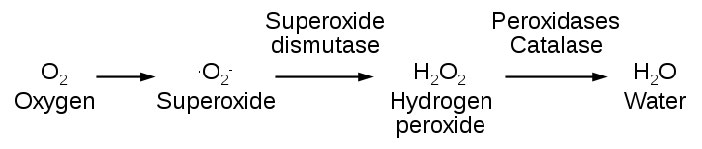
Reactive oxygen species (ROS – Figure 5.37) are oxygen containing molecules, such as peroxides, hydroxyl radical, superoxide, peroxynitrite, and others that are very chemically reactive. Though some ROS, such as peroxide and nitric oxide have important biological functions in signaling, increases in reactive oxygen species in times of stress can cause significant damage in the cell. Exogenous sources of ROS, such as pollution, tobacco, smoke, radiation or drugs can also cause significant problems.
Endogenous production of ROS is directed towards intracellular signaling (H2O2 and nitric oxide, for example) and defense. Many cells, for example, have NADPH oxidase (Figure 5.38) embedded in the exterior portion of the plasma membranes, in peroxisomes, and endoplasmic reticulum. It produces superoxides in the reaction below to kill bacteria .
In the immune system, cells called phagocytes engulf foreign cells and then use ROS to kill them. ROS can serve as signals for action. In zebrafish, damaged tissues have increased levels of H2O2 and this is thought to be a signal for white blood cells to converge on the site. In fish lacking the genes to produce hydrogen peroxide, white blood cells do not converge at the damage site. Sources of hydrogen peroxide include peroxisomes, which generate it as a byproduct of oxidation of long chain fatty acids.
Aging
Reactive oxygen species are at the heart of the free radical theory of aging, which states that organisms age due to the accumulation of damage from free radicals in their cells. In yeast and Drosophila, there is evidence that reducing oxidative damage can increase lifespan. In mice, increasing oxidative damage decreases life span, though in Caenorhabditis, blocking production of superoxide dismutase actually increases lifespan, so the role of ROS in aging is not completely clear.
It is clear, though, that accumulation of mitochondrial damage is problematic for individual cells. Bcl-2 proteins on the surface of mitochondria monitor damage and if they detect it, will activate proteins called Bax to stimulate the release of cytochrome c from the mitochondrial membrane, stimulating apoptosis (programmed cell death). Eventually the dead cell will be phagocytosed.
A common endogenous source of superoxide is the electron transport chain. Superoxide can be produced when movement of electrons into and out of the chain don’t match well. Under these circumstances, semi-reduced CoQ can donate an electron to O2 to form superoxide (O2-). Superoxide can react with many molecules, including DNA where it can cause damage leading to mutation. If it reacts with the aconitase enzyme, ferrous iron (Fe++) can be released which, in turn, can react in the Fenton reaction to produce another reactive oxygen species, the hydroxyl radical (Figure 5.39) .
Countering the effects of ROS are enzymes, such as catalase, superoxide dismutase, and anti-oxidants, such as glutathione and vitamins C and E.
Glutathione protects against oxidative damage by being a substrate for the enzyme glutathione peroxidase. Glutathione peroxidase catalyzes the conversion of hydrogen peroxide to water (next page).
Catalase
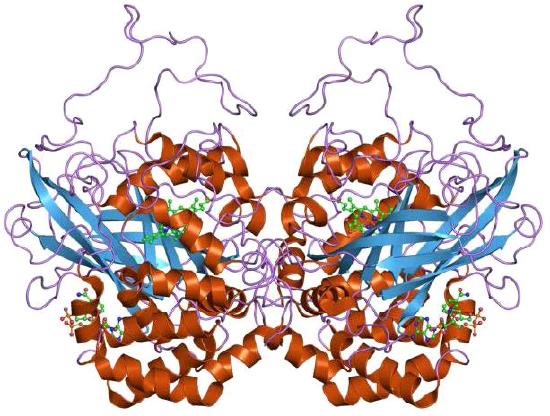
Catalase (Figure 5.40) is an important enzyme for cells of all types that live in an oxygen environment. A first line of defense against reactive oxygen species, catalase catalyzes the breakdown of hydrogen peroxide into water and oxygen.
2 H2O2 <=> 2 H2O + O2
The enzyme, which employs four heme groups in its catalysis, works extremely rapidly, converting up to 40,000,000 molecules of hydrogen peroxide to water and oxygen per enzyme per second. It is abundantly found in peroxisomes.
In addition to catalase’s ability to break down hydrogen peroxide, the enzyme can also use hydrogen peroxide to oxidize a wide variety organic compounds, including phenols, formic acid, formaldehyde, acetaldehyde, and alcohols, but with much lower efficiency.
Health
The importance of catalase for health is uncertain. Mice deficient in the enzyme appear healthy and humans with low levels of the enzyme display few problems. On the other hand, mice engineered to produce higher levels of catalase, in at least one study, lived longer. The ability of organisms to live with lower levels or no catalase may arise from another group of enzymes, the peroxiredoxins, which also act on hydrogen peroxide and may make up for lower quantities of catalase. Last, there is evidence that reduced levels of catalase with aging may be responsible for the graying of hair. Higher levels of H2O2 with reduced catalase results in a bleaching of hair follicles.
Superoxide dismutase
Another important enzyme for protection against reactive oxygen species is superoxide dismutase (SOD), which is found, like catalase, in virtually all organisms living in an oxygen environment. Superoxide dismutase, also like catalase, has a very high Kcat value and, in fact, has the highest Kcat/Km known for any known enzyme. It catalyzes the reactions at the top of the next column (superoxides shown in red):
The enzyme thus works by a ping-pong (double displacement) mechanism (see HERE), being converted between two different forms.
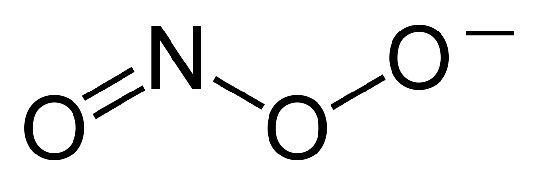
The hydrogen peroxide produced in the second reaction is easily handled by catalase and is also less harmful than superoxide, which can react with nitric oxide (NO) to form very toxic peroxynitrite ions (Figure 5.43). Peroxynitrite has negative effects on cells, as shown in Figure 5.45.
In addition to copper, an ion of Zn++ is also bound by the enzyme and likely plays a role in the catalysis. Forms of superoxide dismutase that use manganese, nickel, or iron are also known and are mostly found in prokaryotes and protists, though a manganese SOD is found in most mitochondria. Copper/zinc enzymes are common in eukaryotes.
Three forms of superoxide dismutase are found in humans and localized to the cytoplasm (SOD1 – Figure 5.45), mitochondria (SOD2 – Figure 5.46), and extracellular areas (SOD3 – Figure 5.47). Mice lacking any of the three forms of the enzyme are more sensitive to drugs, such as paraquat. Deficiency of SOD1 in mice leads to hepatocellular carcinoma and early loss of muscle tissue related to aging. Drosophila lacking SOD2 die before birth and those lacking SOD1 prematurely age.
In humans, superoxide dismutase mutations are associated with the genetically-linked form of Amyotrophic Lateral Sclerosis (ALS) and over-expression of the gene is linked to neural disorders associated with Down syndrome.
Mixed function oxidases
Other enzymes catalyzing reactions involving oxygen include the mixed function oxidases. These enzymes use molecular oxygen for two different purposes in one reaction. The mixed function part of the name is used to indicate reactions in which two different substrates are being oxidized simultaneously. Monooxygenases are examples of mixed function oxidases. An example of a mixed function oxidase reaction is shown below.
AH + BH2 + O2 <=> AOH + B + H2O
In this case, the oxygen molecule has one atom serve as an electron acceptor and the other atom is added to the AH, creating an alcohol.
Cytochrome P450 enzymes
Cytochrome P450 enzymes (called CYPs) are family of heme-containing mixed function oxidase enzymes found in all domains of life. Over 21,000 CYP enzymes are known. The most characteristic reaction catalyzed by these enzymes follows
Monooxygenase reactions such as this are relatively rare in the cell due to their use of molecular oxygen. CYPs require an electron donor for reactions like the one shown here and frequently require a protein to assist in transferring electrons to reduce the heme iron. There are six different classes of P450 enzymes based on how they get electrons
1. Bacterial P450 – electrons from ferredoxin reductase and ferredoxin
2. Mitochondrial P450 – electrons from adrenodoxin reductase and adrenodoxin
3. CYB5R/cyb5 – electrons come from cytochrome b5
4. FMN/Fd – use a fused FMN reductase
5. Microsomal P450 – NADPH electrons come via cytochrome P450 reductase or from cytochrome b5 and cytochrome b5 reductase
6. P450 only systems – do not require external reducing power
The CYP genes are abundant in humans and catalyze thousand of reactions on both cellular and extracellular chemicals. There are 57 human genes categorized into 18 different families of enzymes. Some CYPs are specific for one or a few substrates, but others can act on many different substrates.
CYP enzymes are found in most body tissues and perform important functions in synthesis of steroids (cholesterol, estrogen, testoterone, Vitamin D, e.g.), breakdown of endogenous compounds (bilirubin), and in detoxification of toxic compounds including drugs. Because they act on many drugs, changes in CYP activity can produce unexpected results and cause problems with drug interactions.
Bioactive compounds, for example, in grapefruit juice, can inhibit CYP3A4 activity, leading to increased circulating concentrations of drugs that would normally have been acted upon by CYP3A4. This is the reason that patients prescribed drugs that are known to be CYP3A4 substrates are advised to avoid drinking grapefruit juice while under treatment. St. Johns Wort, an herbal treatment, on the other hand, induces CYP3A4 activity, but inhibits CYP1A1, CYP1B1, and CYP2D6. Tobacco smoke induces CYP1A2 and watercress inhibits CYP2E1.
Cytochromes
Cytochromes are heme-containing proteins that play major roles in the process of electron transport in the mitochondrion and in photosynthesis in the chloroplast. They exist either as monomers (cytochrome c) or as subunits within large redox complexes (Complex III and Complex IV of electron transport. An atom of iron at the center of the heme group plays a central role in the process, flipping between the ferrous (Fe++) and ferric (Fe+++) states as a result of the movement of electrons through it.
There are several different cytochromes. Cytochrome c (Figure 5.47) is a soluble protein loosely associated with the mitochondrion. Cytochromes a and a3 are found in Complex IV. Complex III has cytochromes b and c1 and the plastoquinol-plastocyanin reductase of the chloroplast contains cytochromes b6 and f. Another important class of enzymes containing cytochromes is the cytochrome P450 oxidase group (see above). They get their name from the fact that they absorb light at 450 nm when their heme iron is reduced.
Iron-Sulfur Proteins
Iron-sulfur proteins contain iron-sulfur clusters in a variety of formats, including sulfide-linked di-, tri-, and tetrairon centers existing in different oxidation states (Figures 5.48 & 5.49). The clusters play a variety of roles, but the best known ones are in electron transport where they function in the redox reactions involved in the movement of electrons.
Complexes I and Complex II contain multiple Fe-S centers. Iron-sulfur proteins, though, have many other roles in cells. Aconitase uses an iron-sulfur center in its catalytic action and the ability of the enzyme to bind iron allows it to function as a barometer of iron concentration in cells. Iron-sulfur centers help to generate radicals in enzymes using S-Adenosyl Methionine (SAM) and can serve as a source of sulfur in the synthesis of biotin and lipoic acid. Some iron-sulfur proteins even help to regulate gene expression.
Ferredoxin
Ferredoxins are iron-sulfur containing proteins performing electron transfer in a wide variety of biological systems and processes. They include roles in photosynthesis in chloroplasts. Ferredoxins are classified structurally by the iron-sulfur clustered centers they contain. Fe2S2 clusters (Figure 5.50) are found in chloroplast membranes and can donate electrons to glutamate synthase, nitrate reductase, and sulfite reductase and serve as electron carriers between reductase flavoproteins and bacterial dioxygenase systems. Adrenodoxin is a soluble human Fe2S2 ferredoxin (also called ferredoxin 1) serving as an electron carrier (to cytochrome P450) in mitochondrial monooxygenase systems. Fe4S4 ferredoxins are subdivided as low and high potential ferredoxins, with the latter ones functioning in anaerobic electron transport chains.
Ferritin
Ferritin is an intracellular iron-storage protein found in almost all living organisms, from bacteria to higher plants and animals. It is a globular protein complex with 24 subunits and is the primary intracellular iron-storage protein in eukaryotes and prokaryotes. Ferritin functions to keep iron in a soluble and non-toxic form. Its ability to safely store iron and release it in a controlled fashion allow it to act like the prime iron buffer and solubilizer in cells – keeping the concentration of free iron from going to high or falling too low. Ferritin is located in the cytoplasm in most tissues, but it is also found in the serum acting as an iron carrier. Ferritin that doesn’t contain any iron is known as apoferritin.
Monoamine oxidases
Monoamine oxidases are enzymes that catalyze the oxidative deamination of monoamines, such as serotonin, epinephrine, and dopamine. Removal of the amine with oxygen results in the production of an aldehyde and ammonia. The enzymes are found inside and outside of the central nervous system.
There are two types of monoamine oxidase enzymes – MAO-A and MAO-B. MAO-A is particularly important for oxidizing monoamines consumed in the diet. Both MAO-A and MAO-B play important roles in inactivating monoaminergic neurotransmitters. Both enzymes act on dopamine, tyramine (Figure 5.50), and tryptamine. MAO-A is the primary enzyme for metabolizing melatonin, serotonin, norepinephrine, and epinephrine, while MAO-B is the primary enzyme for phenethylamine (Figure 5.51) and benzylamine. MAO-B levels have been reported to be considerably reduced with tobacco usage.
Actions of monoamine oxidases thus affects levels of neurotransmitters and consequently are thought to play roles in neurological and/or psychiatric disorders. Aberrant levels of MAOs have been linked to numerous psychological problems, including depression, attention deficit disorder (ADD), migraines, schizophrenia, and substance abuse. Medications targeting MAOs are sometimes used to treat depression as a last resort – due to potential side effects. Excess levels of catecholamines, such as epinephrine, norepinephrine, and dopamine, can result in dangerous hypertension events.
DNA damage theory of aging
The DNA Damage Theory of Aging is based on the observation that, over time, cells are subject to extensive oxidative events. As already noted, these afford opportunities for the formation of ROS that can damage cellular molecules, and it follows that accumulation of such damage, especially to the DNA would be deleterious to the cell. The build-up of DNA damage could, thus, be responsible for the changes in gene expression that we associate with aging.
Numerous damage events
The amount of DNA damage that can occur is considerable. In mice, for example, it is estimated that each cell experiences 40,000 to 150,000 damage events per day. The damage, which happens to nuclear as well as to mitochondrial DNA, can result in apoptosis and/or cellular senescence. DNA repair systems, of course, protect against damage to DNA, but over time, unrepairable damage may accumulate.
Oxidative damage
DNA damage can occur in several ways. Oxidation can damage nucleotides and alter their base-pairing tendencies. Oxidation of guanine by reactive oxygen species, for example, can produce 8-oxo-guanine (Figures 5.52 and 5.53). This oxidized nucleobase commonly produced lesion in DNA arising from action of reactive oxygen species like superoxides. 8-oxoguanine is capable of forming a stable base pairing interaction within a DNA duplex with adenine, potentially giving rise to a mutation when DNA replication proceeds. 8-oxoguanine can be repaired if recognized in time by a DNA glycosylase, which acts to clip out the damaged base and it can then be replaced by the proper one. Polycyclic aromatic hydrocarbons from cigarette smoke, diesel exhaust, or overcooked meat can covalently bind to DNA and, if unrepaired, lead to mutation. Chemical damage to DNA can result in broken or cross-linked DNAs.
Diseases of DNA repair
The importance of DNA repair in the aging process is made clear by diseases affecting DNA repair that lead to premature aging. These include Werner syndrome, for whom the life expectancy is 47 years. It arises as a result of loss of two enzymes in base excision repair. People suffering from Cockayne syndrome have a life expectancy of 13 years due to mutations that alter transcription-coupled nucleotide excision repair, which is an important system for fixing oxidative damage.
Further, the life expectancies of 13 species of mammalian organisms correlates with the level of expression of the PARP DNA repair-inducing protein. Interestingly, people who lived past the age of 100 had a higher level of PARP than younger people in the population.
Antioxidants
There is a growing interest in the subject of antioxidants because of health concerns raised by our knowledge of problems created as a result of spontaneous oxidation of biomolecules by Reactive Oxygen Species (ROS), such as superoxide. Antioxidants have the chemical property of protecting against oxidative damage by being readily oxidized themselves, preferentially to other biomolecules.
Biologically, cells have several lines of antioxidant defense. They include molecules, such as vitamins C, A, and E, glutathione, and enzymes that destroy ROS such as superoxide dismutase, catalase, and peroxidases.

Health effects
Oxidation by ROS is mutagenic and has been linked to atherosclerosis. Nonetheless, randomized studies of oral supplementation of various vitamin combinations have shown no protective effect against cancer and supplementation of Vitamin E and selenium has revealed no decrease in the risk of cardiovascular disease. Further, no reduction in mortality rates as a result of supplementation with these materials has been found, so the protective effects, if any, of antioxidants on ROS in human health remain poorly understood.
Glutathione
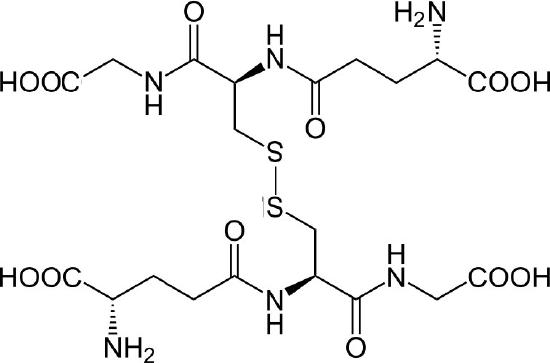
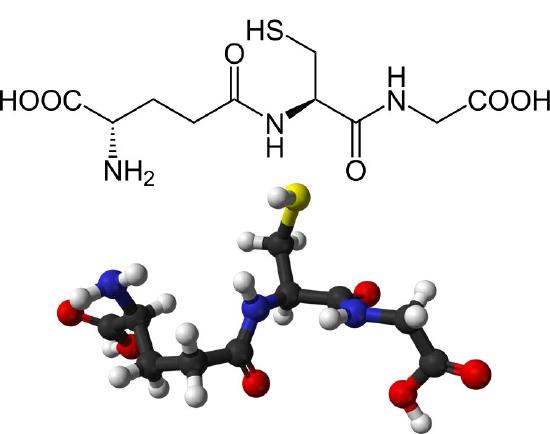
The major endogenous antioxidant found in cells spanning most living systems, glutathione is a tripeptide protecting cells against damage caused by reactive oxygen species and heavy metals (Figures 5.55 & 5.56). The three amino acids in glutathione (glutamate, cysteine, and glycine) are joined in an unusual fashion. The glutamate is joined to the center cysteine by a peptide bond between the R-group carboxyl of glutamate and the α-amine of cysteine. The bond between cysteine and glycine is a normal peptide bond between the α-carboxyl of cysteine and the α-amine of glycine.
The thiol group of cysteine is a reducing agent that reduces disulfide bonds to sulfhydryls in cytoplasmic proteins. This, in turn, is the bridge when two glutathiones get oxidized and form a disulfide bond with each other (Figure 5.56). Glutathione’s two oxidative states are abbreviated as follows: GSH (reduced) and GSSG (oxidized).
Disulfide-joined glutathiones can be separated by reduction of their bonds with glutathione reductase, using electrons from NADPH for the reduction.
Non-ribosomal synthesis
Glutathione is not made by ribosomes. Rather, two enzymes catalyze its synthesis. The enzyme γ-glutamylcysteine synthetase catalyzes the joining of the glutamate to the cysteine and then glutathione synthetase catalyzes the peptide bond formation between the cysteine and the glycine. Each step requires energy from ATP.
Essential for life
Glutathione is important for life. Mice lacking the first enzyme involved in its synthesis in the liver die in the first month after birth. In healthy cells, 90% of glutathione is in the GSH state. Higher levels of GSSG correspond to cells that are oxidatively stressed.
Besides reducing disulfide bonds in cells, glutathione is also important for the following:
- Neutralization of free radicals and reactive oxygen species.
- Maintenance of exogenous antioxidants such as vitamins C and E in their reduced forms.
- Regulation of the nitric oxide cycle
References
1. Winge, D.R., Mol Cell Biol. 2012 Jul; 32(14): 2647–2652. doi: 10.1128/MCB.00573-12
Energy: Electron Transport & Oxidative Phosphorylation
430
Figure 5.14 – Overview of electron transport (bottom left and top right) and oxidative phosphorylation (top left – yellow box) in the mitochondrion
431
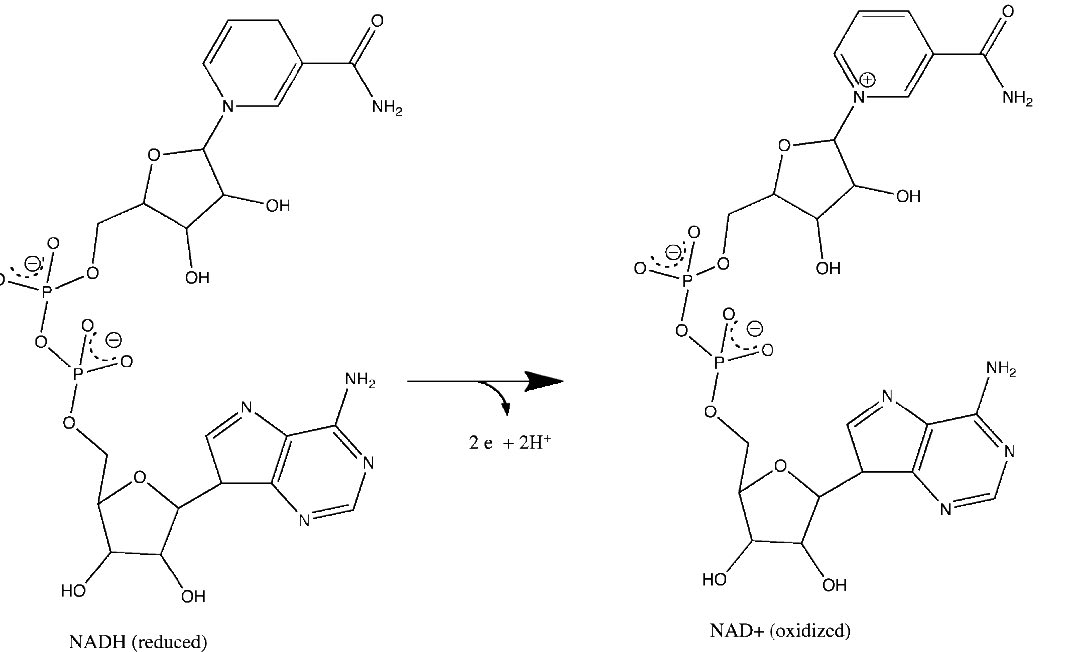
Figure 5.15 – Loss of electrons by NADH to form NAD+. Relevant reactions occur in the top ring of the molecule.
432
Figure 5.16 – Flow of electrons from NADH into the electron transport system. Entry is through complex I
Image by Aleia Kim
Figure 5.17 – Flow of electrons from FADH2 into the electron transport chain. Entry is through complex II.
Image by Aleia Kim
433
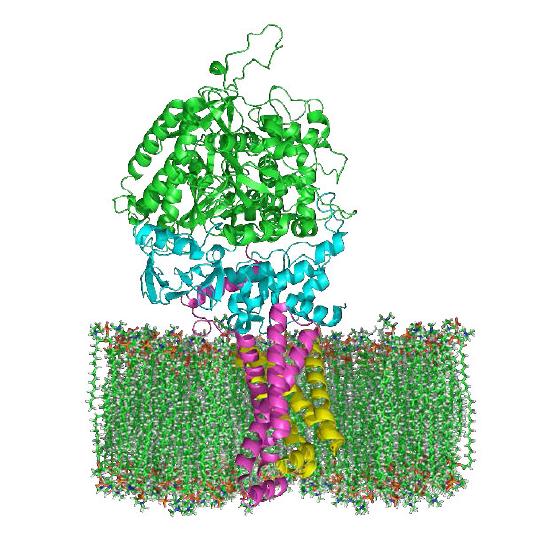
Figure 5.18 – Complex I embedded in the inner mitochondrial membrane. The mitochondrial matrix at at the top
Wikipedia
434
Figure 5.19 – Complex II embedded in inner mitochondrial membrane. Matrix is up.
Wikipedia
435
Figure 5.20 – Movement of electrons through complex I from NADH to coenzyme Q. The mitochondrial matrix is at the bottom
Image by Aleia Kim
Figure 5.21 – Movement of electrons from succinate through complex II (A->B->C->D->Q). Mitochondrial matrix on bottom.
Image by Aleia Kim
436
Figure 5.22 – Complex II in inner mitochondrial membrane showing electron flow. Matrix is up.
Wikipedia
Figure 5.23 – Coenzyme Q
437
Movie 5.2 – The Q-cycle
Wikipedia
Figure 5.24 – The Q-Cycle Image by Aleia Kim
Figure 5.24 – Complex III
Wikipedia
Figure 5.25 – The Q-cycle. Matrix is down.
Image by Aleia Kim
439
Figure 5.26 – Movement of electrons and protons through complex IV. Matrix is down
Image by Aleia Kim
Figure 5.25 – Cytochrome c with bound heme Group
Wikipedia
440
Figure 5.27 – Mitochondrial anatomy. Electron transport complexes and ATP synthase are embedded in the inner mitochondrial membrane
Image by Aleia Kim
441
Figure 5.28 – ATP synthase. Protons pass from intermembrane space (top) through the complex and exit in the matrix (bottom).
Image by Aleia Kim
442
Movie 5.3 – ATP Synthase – ADP + Pi (pink) and ATP (red). The view is end-on from the cytoplasmic side viewing the β subunits
Movie 5.3 – ATP Synthase – ADP + Pi (pink) and ATP (red). The view is end-on from the cytoplasmic side viewing the β subunits
443
Figure 5.29 – Important structural features of the ATP synthase
Image by Aleia Kim
444
Figure 5.30 – Loose (L), Tight (T), and Open (O) structures of the F1 head of ATP synthase. Change of structure occurs by rotation of γ-protein (purple) in center as a result of proton movement. Individual α and β units do not rotate
Image by Aleia Kim
445
Figure 5.31 – Respiration overview in eukaryotic cells
Wikipedia
446
Rest
ATP High / ADP Low
Oxidative Phosphorylation Low
Electron Transport Low
Oxygen Use Low
NADH High / NAD+ Low
Citric Acid Cycle Slow
Exercise
ATP Low / ADP High
Oxidative Phosphorylation High
Electron Transport High
Oxygen Use High
NADH Low / NAD+ High
Citric Acid Cycle Fast
447
Figure 5.32 – Three inhibitors of electron transport
Image by Aleia Kim
448
Figure 5.33 – Oligomycin A – An inhibitor of ATP synthase
Figure 5.34 – 2,4 DNP – an uncoupler of respiratory control
449
In Cells With Tight Coupling
O2 use depends on metabolism
NAD+ levels vary with exercise
Proton gradient high with no exercise
Catabolism depends on energy needs
ETS runs when OxPhos runs and vice versa
In Cells That Are Uncoupled
O2 use high
NAD+ Levels high
Little or no proton gradient
Catabolism high
OxPhos does not run, but ETS runs rapidly
450
451
Figure 5.35 – Alternative oxidase (AOX) of fungi, plants, and protozoa bypasses part of electron transport by taking electrons from CoQ and passing them to oxygen.
452
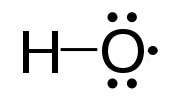
Figure 5.36 – Structure of an oxygen free radical
Wikipedia
NADPH + 2O2
NADP+ + 2O2− + H+
Figure 5.37 – Three sources of reactive oxygen species (ROS) in cells
Wikipedia
453
454
Figure 5.38 A hydroxyl radical
Wikipedia
455
Reduced Glutathione (GSH) + H2O2
Oxidized Glutathione (GSSG) + H2O
Figure 5.40 – Detoxifying reactive oxygen species
Figure 5.39 – Catalase
456
1. O2- + Enzyme-Cu++
O2 + Enzyme-Cu+
2. O2- + Enzyme-Cu+ + 2H+
H2O2 + Enzyme-Cu++
Figure 5.41 – SOD2 of humans
Figure 5.42 3 – Peroxynitrite Ion
Figure 5.44 – SOD1 of humans
Wikipedia
Figure 5.45 – SOD3 of humans
457
Figure 5.43 – Peroxynitrite’s effects on cells lead to necrosis or apoptosis
Wikipedia
458
RH + O2 + NADPH + H+
ROH + H2O + NADP+
459
Figure 5.46 – Cytochrome c with its heme group
460
Figure 5.47 – Fe2S2 Cluster
Figure 5.48 – Redox reactions for Fe4S4 clusters
461
Figure 5.49 – Tyramine
Figure 5.50 – Phenethylamine
462
Figure 5.51 – Guanine and 8-oxo-guanine
Figure 5.52 – Adenine-8-oxo-guanine base pair. dR = deoxyribose
463
Figure 5.53 – Good antioxidant sources
464
Figure 5.55 – Oxidized glutathiones (GSSG) joined by a disulfide bond
Wikipedia
Figure 5.54 – Structure of reduced glutathione (GSH)
465
Figure 5.56 – Resveratrol
466

