DNA Isolation, Gel Electrophoresis, and PCR
To accomplish any of the applications described above, biotechnologists must be able to extract, manipulate, and analyze nucleic acids.
Review of Nucleic Acid Structure
To understand the basic techniques used to work with nucleic acids, remember that nucleic acids are macromolecules made of nucleotides (a sugar, a phosphate, and a nitrogenous base). The phosphate groups on these molecules each have a net negative charge. An entire set of DNA molecules in the nucleus of eukaryotic organisms is called the genome. DNA has two complementary strands linked by hydrogen bonds between the paired bases.
Unlike DNA in eukaryotic cells, RNA molecules leave the nucleus. Messenger RNA (mRNA) is analyzed most frequently because it represents the protein-coding genes that are being expressed in the cell.
Isolation of Nucleic Acids
To study or manipulate nucleic acids, the DNA must first be extracted from cells. Various techniques are used to extract different types of DNA (Figure 2). Most nucleic acid extraction techniques involve steps to break open the cell, and then the use of enzymatic reactions to destroy all undesired macromolecules. Cells are broken open using a detergent solution containing buffering compounds. To prevent degradation and contamination, macromolecules such as proteins and RNA are inactivated using enzymes. The DNA is then brought out of solution using alcohol. The resulting DNA, because it is made up of long polymers, forms a gelatinous mass. This method extracts all the nucleic acid within a cell. This includes genomic DNA (all the DNA in the genome), as well as RNA. If this DNA was to be used for further study, the RNA would often be digested with an enzyme to remove it.
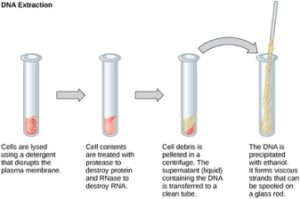
RNA is studied to understand gene expression patterns in cells. RNA is naturally very unstable because enzymes that break down RNA are commonly present in nature. Some are even secreted by our own skin and are very difficult to inactivate. Similar to DNA extraction, RNA extraction involves the use of various buffers and enzymes to inactivate other macromolecules and preserve only the RNA.
Gel Electrophoresis
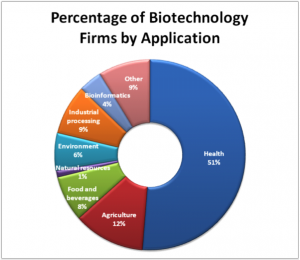
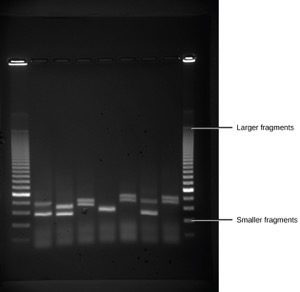
Because nucleic acids are negatively charged ions at neutral or alkaline pH in an aqueous environment, they can be moved by an electric field. Gel electrophoresis is a technique used to separate charged molecules on the basis of size and charge. The nucleic acids can be separated as whole chromosomes or as fragments. The nucleic acids are loaded into a slot at one end of a gel matrix, an electric current is applied, and negatively charged molecules are pulled toward the opposite end of the gel (the end with the positive electrode). Smaller molecules move through the pores in the gel faster than larger molecules; this difference in the rate of migration separates the fragments on the basis of size. The nucleic acids in a gel matrix are invisible until they are stained with a compound that allows them to be seen, such as a dye. Distinct fragments of nucleic acids appear as bands at specific distances from the top of the gel (the negative electrode end) that are based on their size (Figure 3). A mixture of many fragments of varying sizes appear as a long smear, whereas uncut genomic DNA is usually too large to run through the gel and forms a single large band at the top of the gel.
Polymerase Chain Reaction
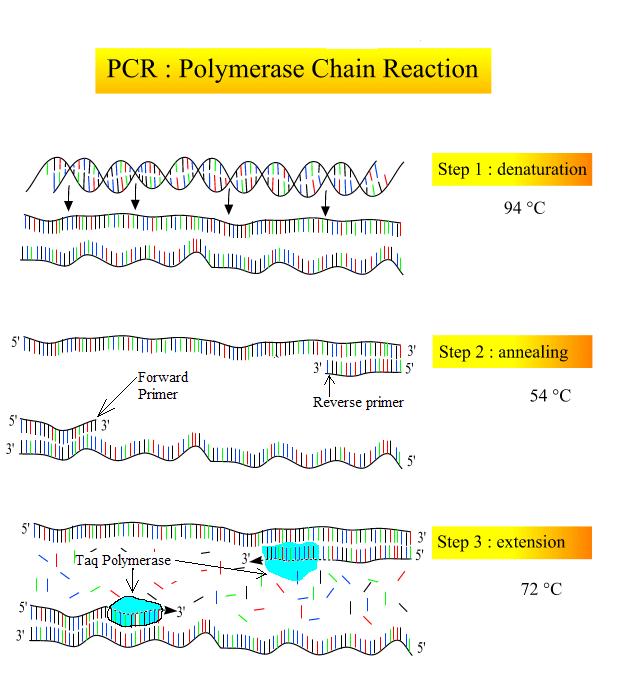
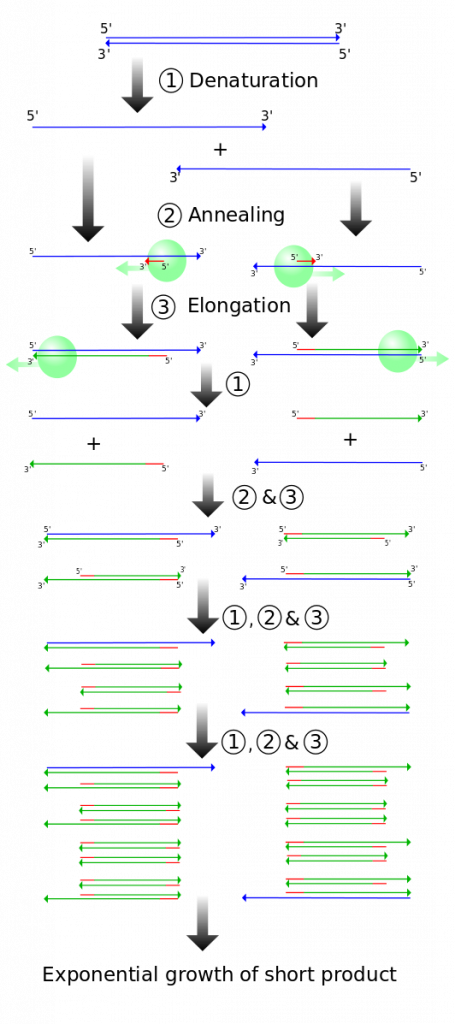
DNA analysis often requires focusing on one or more specific regions of the genome. It also frequently involves situations in which only one or a few copies of a DNA molecule are available for further analysis. These amounts are insufficient for most procedures, such as gel electrophoresis. Polymerase chain reaction (PCR) is a technique used to rapidly increase the number of copies of one specific region of DNA for further analyses (Figure 4). Typically the DNA that is used as the starting sample in a PCR reaction is genomic DNA, which would contain all the genes in the organism. PCR uses a special form of heat tolerant DNA polymerase, the enzyme that replicates DNA, and other short nucleotide sequences called primers that base pair to a specific portion of the DNA being copied. A PCR reaction does not copy the entire genome, rather it makes millions of copies of one specific region of interest.
- Taq DNA polymerase – this polymerase was isolated from Thermus aquaticus, which is a bacteria that lives at high temperatures in hot springs and deep sea vents. Taq polymerase has an optimum temperature of 70-80ºC and can survive nearly an hour at 95ºC. Like other DNA polymerases, Taq polymerase assembles nucleotides only in the 5′ to 3′ direction.
- Primers – Just like during DNA replication, Taq polymerase needs a free 3′ end to begin synthesis of the new DNA. Primers in a PCR reaction are man-made synthetic segments of DNA that match the ends of the sequence that the scientist is interested in amplifying. In order to synthesize both strands of the DNA double helix, a forward and a reverse primer must be added. These two primers have free 3′ ends that point towards the sequence of interest (Figure 4, middle).
PCR is used for many purposes in laboratories. These include: 1) the identification of the owner of a DNA sample left at a crime scene; 2) paternity analysis; 3) the comparison of small amounts of ancient DNA with modern organisms; and 4) determining the sequence of nucleotides in a specific region. In all of these cases, the starting sample is genomic DNA. In some cases, the complete genome may not be present due to the DNA being old or broken down.
PCR involves the following basic steps:
- Denaturation (94ºC): The sample is heated in a small plastic tube inside a thermocycler (which is able to rapidly change temperatures very accurately). Heating the sample to 94ºC causes the hydrogen bonds holding the DNA double helix to denature so the strands are separated (Figure 4, top). Taq polymerase is heat tolerant and does not denature at this temperature.
- Annealing (54-60ºC): The sample is cooled so that the primers can anneal (base pair) with their complementary sequence (Figure 4; middle). Because of the base pairing rules, primers can only anneal in the specific spot that contains their complementary base sequence. This allows scientists to choose which region of DNA will be amplified.
- Extension (72ºC): The sample is warmed up to the optimum temperature for Taq polymerase. Taq extends off of the free 3′ end of both primers, producing a double helix (Figure 4; bottom).
The three basic steps of a PCR reaction are repeated multiple times (“cycles”). Each cycle causes a roughly 2x increase in the number of amplified copies of the specific section of interest (Figure 5). Remember that only one section of DNA is being amplified due to the specificity of the primers.
For the question below, drag TWO primers to the appropriate location where they would anneal. The arrowhead shows the 3′ end of the primer. Keep in mind that Taq DNA polymerase can only extend from the 3′ of the primer.
References
Unless otherwise noted, images on this page are licensed under CC-BY 4.0 by OpenStax.
OpenStax, Biology. OpenStax CNX. May 27, 2016 http://cnx.org/contents/s8Hh0oOc@9.10:8CA_YwJq@3/Cloning-and-Genetic-Engineering

