Eukaryotic RNA Processing
The mRNA transcript is coated in RNA-stabilizing proteins to prevent it from degrading while it is processed and exported out of the nucleus. The three most important steps of pre-mRNA processing are the addition of stabilizing and signaling factors at the 5′ and 3′ ends of the molecule, and the removal of intervening sequences that do not specify the appropriate amino acids. In rare cases, the mRNA transcript can be “edited” after it is transcribed.
5′ Capping
While the pre-mRNA is still being synthesized, a 7-methylguanosine cap is added to the 5′ end of the growing transcript by a phosphate linkage. This 5′ cap protects the nascent mRNA from degradation. In addition, factors involved in protein synthesis recognize the cap to help initiate translation by ribosomes. Structurally, 7-methylguanosine looks like a guanine nucleotide, but with an added methyl group (Figure 1).
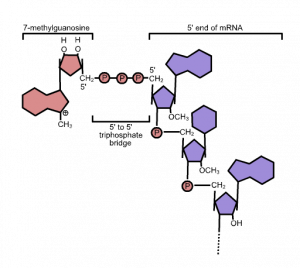
3′ Poly-A Tail
Once elongation is complete, the pre-mRNA is cleaved by an endonuclease between an AAUAAA consensus sequence and a GU-rich sequence, leaving the AAUAAA sequence on the pre-mRNA. An enzyme called poly-A polymerase then adds a string of approximately 200 adenine nucleotides, called the poly-A tail. This modification further protects the pre-mRNA from degradation and signals the export of the cellular factors that the transcript needs to the cytoplasm.
Pre-mRNA Splicing
Eukaryotic genes are composed of exons, which correspond to protein-coding sequences (ex-on signifies that they are expressed), and intervening sequences called introns (int-ron denotes their intervening role), which may be involved in gene regulation but are removed from the pre-mRNA during processing (Figure 2). Intron sequences in mRNA do not encode functional proteins.
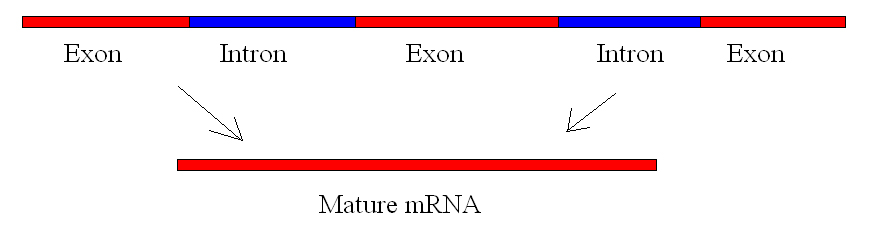
The discovery of introns came as a surprise to researchers in the 1970s who expected that pre-mRNAs would specify protein sequences without further processing, as they had observed in prokaryotes. The genes of higher eukaryotes very often contain one or more introns. These regions may correspond to regulatory sequences; however, the biological significance of having many introns or having very long introns in a gene is unclear. It is possible that introns slow down gene expression because it takes longer to transcribe pre-mRNAs with lots of introns. Alternatively, introns may be nonfunctional sequence remnants left over from the fusion of ancient genes throughout evolution. This is supported by the fact that separate exons often encode separate protein subunits or domains. For the most part, the sequences of introns can be mutated without ultimately affecting the protein product.
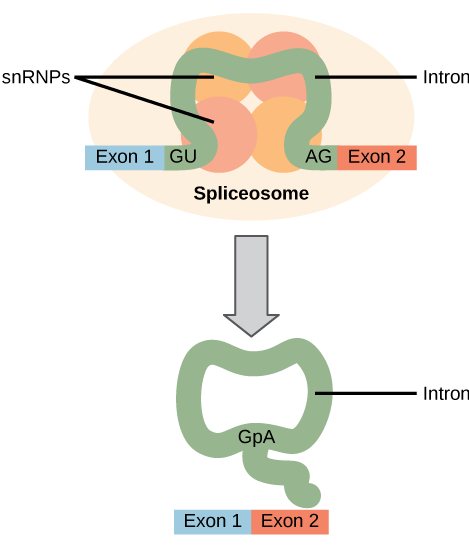
All of a pre-mRNA’s introns must be completely and precisely removed before protein synthesis. If the process errs by even a single nucleotide, the reading frame of the rejoined exons would shift, and the resulting protein would be dysfunctional. The process of removing introns and reconnecting exons is called splicing (Figure 2 and 3). Introns are removed and degraded while the pre-mRNA is still in the nucleus. Splicing occurs by a sequence-specific mechanism that ensures introns will be removed and exons rejoined with the accuracy and precision of a single nucleotide. The splicing of pre-mRNAs is conducted by complexes of proteins and RNA molecules called spliceosomes (Figure 3).
Note that more than 70 individual introns can be present, and each has to undergo the process of splicing—in addition to 5′ capping and the addition of a poly-A tail—just to generate a single, translatable mRNA molecule.
References
Unless otherwise noted, images on this page are licensed under CC-BY 4.0 by OpenStax.
OpenStax, Biology. OpenStax CNX. May 27, 2016 http://cnx.org/contents/s8Hh0oOc@9.10:TkuNUJis@3/Transcription

