Translation
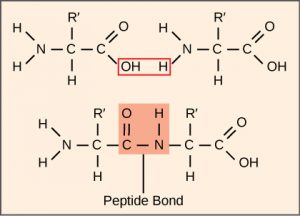
Remember that there are 20 different amino acids that are commonly used. In Figure 1, R’ represents the part of the amino acid which is different between these 20 structures.
The Protein Synthesis Machinery
In addition to the mRNA template, many other molecules contribute to the process of translation and the general structures and functions of the protein synthesis machinery are comparable from bacteria to human cells. Translation requires the input of an mRNA template, ribosomes, tRNAs, and various enzymatic factors (Figure 1).
Ribosomes
Even before an mRNA is translated, a cell must invest energy to build each of its ribosomes. Ribosomes are the part of the cell which reads the information in the mRNA molecule and joins amino acids together in the correct order. A ribosome is a very large, complex macromolecule composed of structural and catalytic rRNAs, and many distinct polypeptides. In eukaryotes, the nucleolus is completely specialized for the synthesis and assembly of rRNAs (the RNA component that makes up ribosomes).
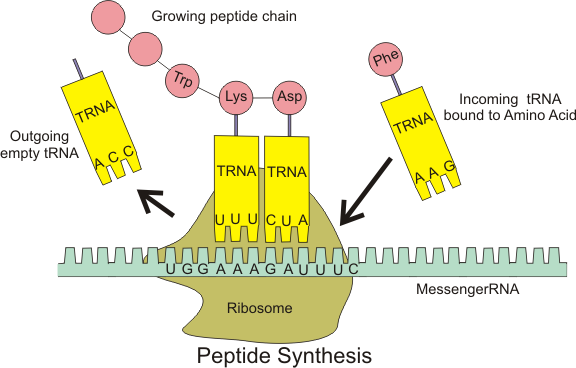
Ribosomes are made up of two subunits that come together for translation, rather like a hamburger bun comes together around the meat (the mRNA). The small subunit is responsible for binding the mRNA template, whereas the large subunit sequentially binds tRNAs, a type of RNA molecule that brings amino acids to the growing chain of the polypeptide. Each mRNA molecule can be simultaneously translated by many ribosomes, all synthesizing protein in the same direction: reading the mRNA from 5′ to 3′ and synthesizing the polypeptide from the N terminus to the C terminus (refer to Figure 1 – the N terminus is the end of the amino acid with the Nitrogen; the C terminus is the end with the Carbon).
Ribosomes exist in the cytoplasm in prokaryotes and in the cytoplasm and rough endoplasmic reticulum in eukaryotes. Mitochondria and chloroplasts also have their own ribosomes in the matrix and stroma, which look more similar to prokaryotic ribosomes (and have similar drug sensitivities) than the ribosomes just outside their outer membranes in the cytoplasm.
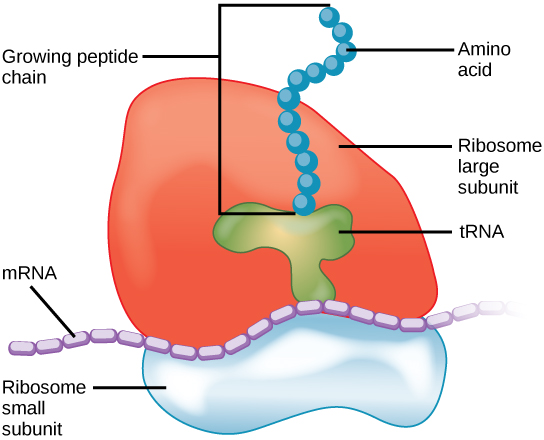
Figure 2 The protein synthesis machinery includes the large and small subunits of the ribosome, mRNA, and tRNA.
tRNA
Depending on the species, 40 to 60 types of tRNA exist in the cytoplasm. Serving as adaptors, specific tRNAs bind to sequences on the mRNA template and add the corresponding amino acid to the polypeptide chain. Therefore, tRNAs are the molecules that actually “translate” the language of RNA into the language of proteins.
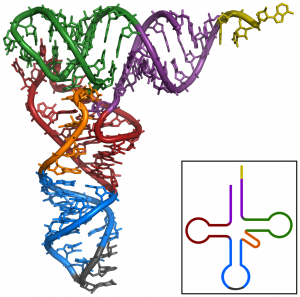
Each tRNA is made up of a linear RNA molecule that is folded into a complex shape (Figure 3). At one end of the tRNA is an anticodon, which recognizes and base pairs with one of the mRNA codons. At the other end, a specific amino acid is attached. Of the 64 possible mRNA codons—or triplet combinations of A, U, G, and C—three specify the termination of protein synthesis and 61 specify the addition of amino acids to the polypeptide chain. Of these 61, one codon (AUG) also encodes the initiation of translation. Each tRNA anticodon can base pair with one of the mRNA codons and add an amino acid or terminate translation, according to the genetic code. For instance, if the sequence CUA occurred on an mRNA template in the proper reading frame, it would bind a tRNA expressing the complementary sequence, GAU, which would be linked to the amino acid leucine.
Aminoacyl tRNA Synthetases
For each tRNA to function, it must have its specific amino acid bonded to it. In the process of tRNA “charging,” each tRNA molecule is bonded to its correct amino acid by a group of enzymes called aminoacyl tRNA synthetases. At least one type of aminoacyl tRNA synthetase exists for each of the 20 amino acids; the exact number of aminoacyl tRNA synthetases varies by species. These enzymes utilize the energy from ATP to energize a specific amino acid, which is then transferred to the tRNA. In this way, tRNA molecules can be used over and over again, but each tRNA always carries the same amino acid because of the specificity of the aminoacyl tRNA synthetase enzymes.
The Mechanism of Protein Synthesis
As with mRNA synthesis, protein synthesis can be divided into three phases: initiation, elongation, and termination. The process of translation is similar in prokaryotes and eukaryotes. Here we’ll explore how translation occurs in E. coli, a representative prokaryote, and specify any differences between prokaryotic and eukaryotic translation.
Initiation of Translation
Protein synthesis begins with the formation of an initiation complex. In E. coli, this complex involves the small ribosomal subunit, the mRNA template, three initiation factors (IF-1, IF-2, and IF-3), and a special initiator tRNA, called tRNAFMet. The initiator tRNA interacts with the start codon AUG, links to a formylated methionine amino acid called fMet, and can also bind IF-2. Formylated methionine is inserted by fMet-tRNAFMet at the beginning of every polypeptide chain synthesized by E. coli, but it is usually clipped off after translation is complete. When an in-frame AUG is encountered during translation elongation, a non-formylated methionine is inserted by a regular Met-tRNAMet.
In E. coli mRNA, a sequence upstream of the first AUG codon, called the Shine-Dalgarno sequence (AGGAGG), interacts with the rRNA molecules that compose the ribosome. This interaction anchors the small ribosomal subunit at the correct location on the mRNA template. Guanosine triphosphate (GTP), which is a purine nucleotide triphosphate, acts as an energy source during translation—both at the start of elongation and during the ribosome’s translocation.
In eukaryotes, a similar initiation complex forms, comprising mRNA, the small ribosomal subunit, IFs, and nucleoside triphosphates (GTP and ATP). The charged initiator tRNA, called Met-tRNAi, does not bind fMet in eukaryotes, but is distinct from other Met-tRNAs in that it can bind IFs. Like in E. coli, a “normal” methionine amino acid is inserted when the ribosome encounters in-frame AUG codons.
Instead of depositing at the Shine-Dalgarno sequence, the eukaryotic initiation complex recognizes the 7-methylguanosine cap at the 5′ end of the mRNA. A cap-binding protein (CBP) and several other IFs assist the movement of the ribosome to the 5′ cap. Once at the cap, the initiation complex tracks along the mRNA in the 5′ to 3′ direction, searching for the AUG start codon. Many eukaryotic mRNAs are translated from the first AUG, but this is not always the case. The sequence of bases around each AUG helps determine if that AUG will be used as the start codon.
Once the appropriate AUG is identified, the large ribosomal subunit binds to the complex of Met-tRNAi, mRNA, and the small ribosomal subunit. This step completes the initiation of translation in eukaryotes.
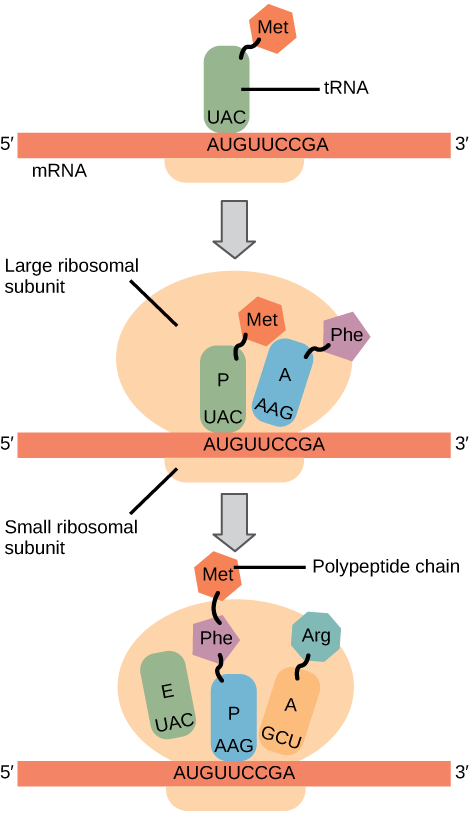
Summary: In both prokaryotes and eukaryotes, the small ribosomal subunit binds to the special initiator methionine tRNA. With the help of several other factors, this complex identifies the start codon (AUG) based on the sequence of nucleotides nearby (Figure 4, top diagram). Then, the large ribosomal subunit binds (Figure 4, middle diagram).
Translation, Elongation, and Termination
In prokaryotes and eukaryotes, the basics of elongation are the same. The large ribosomal subunit of consists of three compartments: the A (aminoacyl) site binds incoming charged aminoacyl tRNAs. The P (peptidyl) site binds charged tRNAs carrying amino acids that have formed peptide bonds with the growing polypeptide chain but have not yet dissociated from their corresponding tRNA. The E (exit) site releases dissociated tRNAs so that they can be recharged with free amino acids. There is one exception to this assembly line of tRNAs: in E. coli, fMet-tRNAFMet is capable of entering the P site directly without first entering the A site. Similarly, the eukaryotic Met-tRNAi, with help from other proteins of the initiation complex, binds directly to the P site. In both cases, this creates an initiation complex with a free A site ready to accept the tRNA corresponding to the first codon after the AUG.
During translation elongation, the mRNA template provides specificity. As the ribosome moves along the mRNA, each mRNA codon comes into register, and specific binding with the corresponding charged tRNA anticodon is ensured. If mRNA were not present in the elongation complex, the ribosome would bind tRNAs nonspecifically and a nonsense protein would be produced.
Elongation proceeds with charged tRNAs entering the A site and then shifting to the P site followed by the E site with each single-codon “step” of the ribosome (Figure 4, bottom diagram). Ribosomal steps are induced by conformational changes that advance the ribosome by three bases in the 3′ direction. The energy for each step of the ribosome is donated by an elongation factor that hydrolyzes GTP. Peptide bonds form between the amino group of the amino acid attached to the A-site tRNA and the carboxyl group of the amino acid attached to the P-site tRNA. The formation of each peptide bond is catalyzed by peptidyl transferase, an RNA-based enzyme that is integrated into the large ribosomal subunit. The energy for each peptide bond formation is derived from GTP hydrolysis, which is catalyzed by a separate elongation factor. The amino acid bound to the P-site tRNA is also linked to the growing polypeptide chain. As the ribosome steps across the mRNA, the former P-site tRNA enters the E site, detaches from the amino acid, and is expelled (Figure 4). Amazingly, the E. coli translation apparatus takes only 0.05 seconds to add each amino acid, meaning that a 200-amino acid protein can be translated in just 10 seconds.
References
Unless otherwise noted, images on this page are licensed under CC-BY 4.0 by OpenStax.
OpenStax, Biology. OpenStax CNX. January 2, 2017 http://cnx.org/contents/s8Hh0oOc@9.10:FUH9XUkW@6/Translation

