DNA Replication in Prokaryotes
The prokaryotic chromosome is a circular molecule with a less extensive coiling structure than eukaryotic chromosomes. The eukaryotic chromosome is linear and highly coiled around proteins. While there are many similarities in the DNA replication process, these structural differences necessitate some differences in the DNA replication process in these two life forms. DNA replication in prokaryotes has been extensively studied, so we will learn the basic process of prokaryotic DNA replication, then focus on the differences between prokaryotes and eukaryotes.
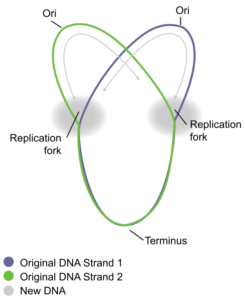
How does the replication machinery know where to start? It turns out that there are specific nucleotide sequences called origins of replication where replication begins. E. coli has a single origin of replication on its one chromosome, as do most prokaryotes (Figure 1). The origin of replication is approximately 245 base pairs long and is rich in AT sequences. This sequence of base pairs is recognized by certain proteins that bind to this site. An enzyme called helicase unwinds the DNA by breaking the hydrogen bonds between the nitrogenous base pairs. ATP hydrolysis is required for this process because it requires energy. As the DNA opens up, Y-shaped structures called replication forks are formed (Figure 1). Two replication forks are formed at the origin of replication and these get extended bi-directionally as replication proceeds. Single-strand binding proteins (Figure 2) coat the single strands of DNA near the replication fork to prevent the single-stranded DNA from winding back into a double helix.
The next important enzyme is DNA polymerase III, also known as DNA pol III, which adds nucleotides one by one to the growing DNA chain (Figure 2). The addition of nucleotides requires energy; this energy is obtained from the nucleotides that have three phosphates attached to them. ATP structurally is an adenine nucleotide which has three phosphate groups attached; breaking off the third phosphate releases energy. In addition to ATP, there are also TTP, CTP, and GTP. Each of these is made up of the corresponding nucleotide with three phosphates attached. When the bond between the phosphates is broken, the energy released is used to form the phosphodiester bond between the incoming nucleotide and the existing chain.
In prokaryotes, three main types of polymerases are known: DNA pol I, DNA pol II, and DNA pol III. DNA pol III is the enzyme required for DNA synthesis; DNA pol I is used later in the process and DNA pol II is used primarily required for repair (this is another irritating example of naming that was done based on the order of discovery rather than an order that makes sense).
DNA polymerase is able to add nucleotides only in the 5′ to 3′ direction (a new DNA strand can be only extended in this direction). It requires a free 3′-OH group (located on the sugar) to which it can add the next nucleotide by forming a phosphodiester bond between the 3′-OH end and the 5′ phosphate of the next nucleotide. This essentially means that it cannot add nucleotides if a free 3′-OH group is not available. Then how does it add the first nucleotide? The problem is solved with the help of a primer that provides the free 3′-OH end. Another enzyme, RNA primase, synthesizes an RNA primer that is about five to ten nucleotides long and complementary to the DNA. RNA primase does not require a free 3′-OH group. Because this sequence primes the DNA synthesis, it is appropriately called the primer. DNA polymerase can now extend this RNA primer, adding nucleotides one by one that are complementary to the template strand (Figure 2).
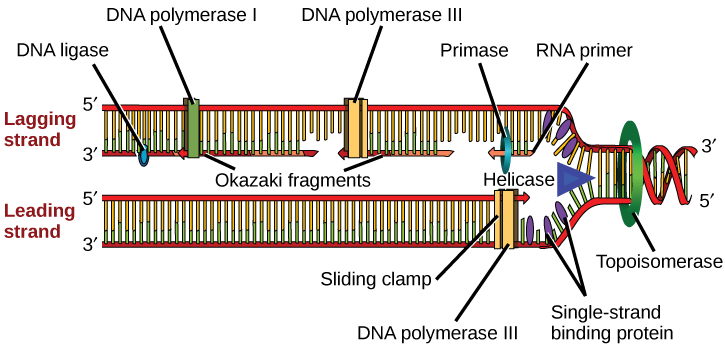
The replication fork moves at the rate of 1000 nucleotides per second. DNA polymerase can only extend in the 5′ to 3′ direction, which poses a slight problem at the replication fork. As we know, the DNA double helix is anti-parallel; that is, one strand is in the 5′ to 3′ direction and the other is oriented in the 3′ to 5′ direction. One strand, which is complementary to the 3′ to 5′ parental DNA strand, is synthesized continuously towards the replication fork because the polymerase can add nucleotides in this direction. This continuously synthesized strand is known as the leading strand. The other strand, complementary to the 5′ to 3′ parental DNA, is extended away from the replication fork, in small fragments known as Okazaki fragments, each requiring a primer to start the synthesis. Okazaki fragments are named after the Japanese scientist who first discovered them. The strand with the Okazaki fragments is known as the lagging strand.
The leading strand can be extended by one primer alone, whereas the lagging strand needs a new primer for each of the short Okazaki fragments. The overall direction of the lagging strand will be 3′ to 5′, and that of the leading strand 5′ to 3′. A protein called the sliding clamp holds the DNA polymerase in place as it continues to add nucleotides. The sliding clamp is a ring-shaped protein that binds to the DNA and holds the polymerase in place. Topoisomerase prevents the over-winding of the DNA double helix ahead of the replication fork as the DNA is opening up; it does so by causing temporary nicks in the DNA helix and then resealing it. As synthesis proceeds, the RNA primers are replaced by DNA pol I, which breaks down the RNA and fills the gaps with DNA nucleotides. The nicks that remain between the newly synthesized DNA (that replaced the RNA primer) and the previously synthesized DNA are sealed by the enzyme DNA ligase that catalyzes the formation of phosphodiester linkage between the 3′-OH end of one nucleotide and the 5′ phosphate end of the other fragment.
This process is hard to visualize from reading the text. Here are three animations / videos to help you think about the process:
- Professor Creech’s DNA Replication drawing lecture: https://www.youtube.com/watch?v=x7hzYGIqV-4&list=PLoRv4Uwz6oYAttlHYPiH4cI3NxKS8YzXP&index=3
- Bozeman Science DNA Replication lecture: https://www.youtube.com/watch?v=FBmO_rmXxIw
- Oxford University Press DNA Replication animation: https://www.youtube.com/watch?v=0Ha9nppnwOc
Once the chromosome has been completely replicated, the two DNA copies move into two different cells during cell division. The process of DNA replication can be summarized as follows:
- DNA unwinds at the origin of replication.
- Helicase opens up the DNA-forming replication forks; these are extended in both directions.
- Single-strand binding proteins coat the DNA around the replication fork to prevent rewinding of the DNA.
- Topoisomerase binds at the region ahead of the replication fork to prevent supercoiling (over-winding).
- Primase synthesizes RNA primers complementary to the DNA strand.
- DNA polymerase III starts adding nucleotides to the 3′-OH (sugar) end of the primer.
- Elongation of both the lagging and the leading strand continues.
- RNA primers are removed and gaps are filled with DNA by DNA pol I.
- The gaps between the DNA fragments are sealed by DNA ligase.
Table 1: The enzymes involved in prokaryotic DNA replication and the functions of each.
| Prokaryotic DNA Replication: Enzymes and Their Function | |
|---|---|
| Enzyme/protein | Specific Function |
| DNA pol I | Exonuclease activity removes RNA primer and replaces with newly synthesized DNA |
| DNA pol II | Repair function |
| DNA pol III | Main enzyme that adds nucleotides in the 5′-3′ direction |
| Helicase | Opens the DNA helix by breaking hydrogen bonds between the nitrogenous bases |
| Ligase | Seals the gaps between the Okazaki fragments to create one continuous DNA strand |
| Primase | Synthesizes RNA primers needed to start replication |
| Sliding Clamp | Helps to hold the DNA polymerase in place when nucleotides are being added |
| Topoisomerase | Helps relieve the stress on DNA when unwinding by causing breaks and then resealing the DNA |
| Single-strand binding proteins (SSB) | Binds to single-stranded DNA to avoid DNA rewinding back. |
DNA replication has been extremely well-studied in prokaryotes, primarily because of the small size of the genome and large number of variants available. Escherichia coli has 4.6 million base pairs in a single circular chromosome, and all of it gets replicated in approximately 42 minutes, starting from a single origin of replication and proceeding around the chromosome in both directions. This means that approximately 1000 nucleotides are added per second. The process is much more rapid than in eukaryotes.
References
Unless otherwise noted, images on this page are licensed under CC-BY 4.0 by OpenStax.
OpenStax, Concepts of Biology. OpenStax CNX. May 18, 2016 http://cnx.org/contents/s8Hh0oOc@9.10:2ousESf0@5/DNA-Replication

