Eukaryotic Translational and Post-Translational Regulation
After the RNA has been transported to the cytoplasm, it is translated into protein. Control of this process is largely dependent on the RNA molecule. As previously discussed, the stability of the RNA will have a large impact on its translation into a protein. As the stability changes, the amount of time that it is available for translation also changes.
The Initiation Complex and Translation Rate
Like transcription, translation is controlled by proteins that bind and initiate the process. In translation, the complex that assembles to start the process is referred to as the initiation complex. Regulation of the formation of this complex can increase or decrease rates of translation (Figure 1).
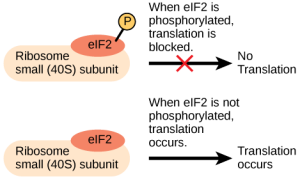
Chemical Modifications, Protein Activity, and Longevity
Proteins can be chemically modified with the addition of groups including methyl, phosphate, acetyl, and ubiquitin groups. The addition or removal of these groups from proteins regulates their activity or the length of time they exist in the cell. Sometimes these modifications can regulate where a protein is found in the cell—for example, in the nucleus, the cytoplasm, or attached to the plasma membrane.
Chemical modifications occur in response to external stimuli such as stress, the lack of nutrients, heat, or ultraviolet light exposure. These changes can alter epigenetic accessibility, transcription, mRNA stability, or translation—all resulting in changes in expression of various genes. This is an efficient way for the cell to rapidly change the levels of specific proteins in response to the environment. Because proteins are involved in every stage of gene regulation, the phosphorylation of a protein (depending on the protein that is modified) can alter accessibility to the chromosome, can alter translation (by altering transcription factor binding or function), can change nuclear shuttling (by influencing modifications to the nuclear pore complex), can alter RNA stability (by binding or not binding to the RNA to regulate its stability), can modify translation (increase or decrease), or can change post-translational modifications (add or remove phosphates or other chemical modifications).
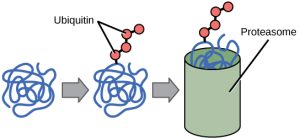
The addition of an ubiquitin group to a protein marks that protein for degradation. Ubiquitin acts like a flag indicating that the protein lifespan is complete. These proteins are moved to the proteasome, an organelle that functions to remove proteins, to be degraded (Figure 2). One way to control gene expression, therefore, is to alter the longevity of the protein.
References
Unless otherwise noted, images on this page are licensed under CC-BY 4.0 by OpenStax.
OpenStax, Concepts of Biology. OpenStax CNX. May 18, 2016 http://cnx.org/contents/b3c1e1d2-839c-42b0-a314-e119a8aafbdd@9.10

