DNA organization inside a cell
DNA Organization in Prokaryotes
A cell’s DNA, packaged as a double-stranded DNA molecule, is called its genome. In prokaryotes, the genome is composed of a single, double-stranded DNA molecule in the form of a loop or circle (Figure 1). The region in the cell containing this genetic material is called a nucleoid (remember that prokaryotes do not have a separate membrane-bound nucleus). Some prokaryotes also have smaller loops of DNA called plasmids that are not essential for normal growth. Bacteria can exchange these plasmids with other bacteria, sometimes receiving beneficial new genes that the recipient can add to their chromosomal DNA. Antibiotic resistance is one trait that often spreads through a bacterial colony through plasmid exchange.
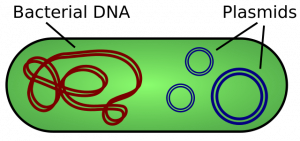
The size of the genome in one of the most well-studied prokaryotes, E.coli, is 4.6 million base pairs (which would be approximately 1.1 mm in length, if cut and stretched out). So how does this fit inside a small bacterial cell? The DNA is twisted by what is known as supercoiling. Supercoiled DNA is coiled more tightly than would be typically be found in a cell (more than 10 nucleotides per twist of the helix). If you visualize twisting a rope until it twists back on itself, you have a pretty good visual of supercoiled DNA. This process allows the DNA to be compacted into the small space inside a bacteria.
DNA Organization in Eukaryotes
Eukaryotes have much more DNA than prokaryotes. For example, an E. coli bacteria contains roughly 3 million base pairs of DNA, while a human contains roughly 3 billion. In eukaryotes such as humans and other animals, the genome consists of several double-stranded linear DNA molecules (Figure 2), which are located inside a membrane-bound nucleus. Each species of eukaryotes has a characteristic number of chromosomes in the nuclei (plural of nucleus) of its cells. A normal human gamete (sperm or egg) contains 23 chromosomes. A normal human body cell, or somatic cell, contains 46 chromosomes (one set of 23 from the egg and one set of 23 from the sperm; Figure 2). The letter n is used to represent a single set of chromosomes; therefore, a gamete (sperm or egg) is designated 1n, and is called a haploid cell. Somatic cells (body cells) are designated 2n and are called diploid cells.
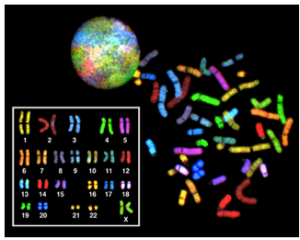
Matched pairs of chromosomes in a diploid organism are called homologous (“same knowledge”) chromosomes. Of a pair of homologous chromosomes, one came from the egg and the second came from the sperm. Homologous chromosomes are the same length and have specific nucleotide segments called genes in exactly the same location, or locus. Genes, the functional units of chromosomes, determine specific characteristics by coding for specific proteins. Traits are the variations of those characteristics. For example, hair color is a characteristic with traits that are blonde, brown, or black.
Each copy of a homologous pair of chromosomes originates from a different parent; therefore, the sequence of DNA present in the two genes on a pair of homologous chromosomes is not necessarily identical, despite the same genes being present in the same locations on the chromosome. These different versions of a gene that contain different sequences of DNA are called alleles.
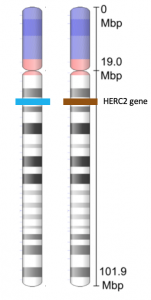
If you look at Figure 3, you can see a pair of homologous chromosomes. The chromosome shown in the figure is chromosome 15. The HERC2 gene is located on this chromosome. The HERC2 gene is one of at least three genes involved in determining eye color. The HERC2 gene is also involved in DNA repair, centrosome assembly, iron metabolism, and is associated with some genetic diseases (Sánchez-Tena et al, 2016). Each person inherits two copies of the HERC2 gene: one from the egg and one from the sperm. However, the alleles of the HERC2 gene that they inherit can be different. In the figure, the cell containing this homologous pair of chromosomes contains one blue allele and one brown allele.
The variation of individuals within a species is due to the specific combination of the genes inherited from both parents. Even a slightly altered sequence of nucleotides within a gene can result in an alternative trait. For example, there are three possible gene sequences (alleles) on the human chromosome that code for blood type: sequence A, sequence B, and sequence O. Because all diploid human cells have two copies of the chromosome that determines blood type, the blood type (the trait) is determined by which two versions of the marker gene are inherited. It is possible to have two copies of the same allele on both homologous chromosomes, with one on each (for example, AA, BB, or OO), or two different alleles, such as AB.
Minor variations of traits, such as blood type, eye color, and handedness, contribute to the natural variation found within a species. However, if the entire DNA sequence from any pair of human homologous chromosomes is compared, the difference is less than one percent. The sex chromosomes, X and Y, are the single exception to the rule of homologous chromosome uniformity: Other than a small amount of homology that is necessary to accurately produce gametes, the genes found on the X and Y chromosomes are different.
Eukaryotic Chromosomal Structure and Compaction
If the DNA from all 46 chromosomes in a human cell nucleus was laid out end to end, it would measure approximately two meters; however, its diameter would be only 2 nm. Considering that the size of a typical human cell is about 10 µm (100,000 cells lined up to equal one meter), DNA must be tightly packaged to fit in the cell’s nucleus. At the same time, it must also be readily accessible for the genes to be expressed. During some stages of the cell cycle, the long strands of DNA are condensed into compact chromosomes.
Eukaryotes, whose chromosomes each consist of a linear DNA molecule, employ a complex type of packing strategy to fit their DNA inside the nucleus (Figure 4). At the most basic level, DNA is wrapped around proteins known as histones to form structures called nucleosomes. The histones are evolutionarily conserved proteins that form an octamer of eight histone proteins attached together. DNA, which is negatively charged because of the phosphate groups, is wrapped tightly around the histone core, which has an overall positive charge. This nucleosome is linked to the next one with the help of a linker DNA. This is also known as the “beads on a string” structure. This is further compacted into a 30 nm fiber, which is the diameter of the structure. At the metaphase stage, the chromosomes are at their most compact, are approximately 700 nm in width, and are found in association with scaffold proteins.
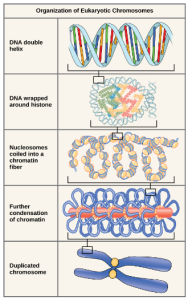
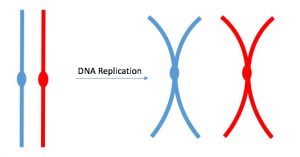
DNA replicates in the S phase of interphase. After replication, the chromosomes are composed of two linked sister chromatids (Figure 5). This means that the only time chromosomes look like an “X” is after DNA replication has taken place and the chromosomes have condensed. During the majority of the cell’s life, chromosomes are composed of only one copy and they are not tightly compacted into chromosomes. When fully compact, the pairs of identically packed chromosomes are bound to each other by cohesin proteins. The connection between the sister chromatids is closest in a region called the centromere. The conjoined sister chromatids, with a diameter of about 1 µm, are visible under a light microscope. The centromeric region is highly condensed and thus will appear as a constricted area. In Figure 4, it is shown as an oval because it is easier to draw that way.
References
Unless otherwise noted, images on this page are licensed under CC-BY 4.0 by OpenStax.
OpenStax, Biology. OpenStax CNX. December 21, 2017 https://cnx.org/contents/GFy_h8cu@10.120:U7tPDRxK@9/DNA-Structure-and-Sequencing
Sánchez-Tena, S., Cubillos-Rojas, M., Schneider, T., Rosa, J.L. 2016. Functional and pathological relevance of HERC family proteins: a decade later. Cell Mol Life Sci. 2016 May;73(10):1955-68. doi: 10.1007/s00018-016-2139-8.

