DNA Replication in Eukaryotes
The essential steps of replication are the same as in prokaryotes. Starting replication is more complex in eukaryotes. At the origin of replication, a pre-replication complex is made with other initiator proteins. Other proteins are then recruited to start the replication process. The overall process is the same, although differently named enzymes fulfill the same function. For example, DNA pol III is used for the majority of replication in prokaryotes, while in eukaryotes the leading strand is continuously synthesized by the enzyme pol δ, the lagging strand is synthesized by pol ε. We are focusing on the enzymes used in prokaryotic replication, so don’t worry about these name differences.
Here are the important differences between prokaryotic and eukaryotic replication:
Table 1: Differences between prokaryotic and eukaryotic replication
| Property | Prokaryotes | Eukaryotes |
|---|---|---|
| Origin of replication | Single | Multiple |
| Rate of replication | 1000 nucleotides/s | 50 to 100 nucleotides/s |
| DNA polymerase types | 5 | 14 |
| DNA packaging | supercoiling | wound around histones |
| Telomerase | Not present | Present |
Origins and rate of replication
Eukaryotic genomes are much more complex and larger in size than prokaryotic genomes. The human genome has three billion base pairs per haploid set of chromosomes, and 6 billion base pairs are replicated during the S phase of the cell cycle. This means that there must be multiple origins of replication on the eukaryotic chromosome in order for all the DNA to be replicated in a timely manner; humans can have up to 100,000 origins of replication. The rate of replication is approximately 100 nucleotides per second, much slower than prokaryotic replication.
DNA polymerase types
The number of DNA polymerases in eukaryotes is much more than prokaryotes: 14 are known, of which five are known to have major roles during replication and have been well studied. They are known as pol α, pol β, pol γ, pol δ, and pol ε. I won’t ever ask you the names of these polymerases – learn the names of the prokaryotic polymerases.
DNA packaging
Eukaryotic DNA is wound around proteins known as histones to form structures called nucleosomes. The DNA must be made accessible in order for DNA replication to proceed. The chromatin (the complex between DNA and proteins) may undergo some chemical modifications, so that the DNA may be able to slide off the histones or otherwise be accessible to the enzymes of the DNA replication machinery. Prokaryotes do not packaged their DNA by wrapping it around histones.
Telomere replication
Unlike prokaryotic chromosomes, eukaryotic chromosomes are linear. As you’ve learned, the enzyme DNA pol can add nucleotides only in the 5′ to 3′ direction. In the leading strand, synthesis continues until the end of the chromosome is reached. On the lagging strand, DNA is synthesized in short stretches, each of which is initiated by a separate primer. When the replication fork reaches the end of the linear chromosome, there is no place for a primer to be made for the DNA fragment to be copied at the end of the chromosome. These ends thus remain unpaired, and over time these ends may get progressively shorter as cells continue to divide.
The ends of the linear chromosomes are known as telomeres, which have repetitive sequences that do not code for a particular gene. These telomeres protect the genes that are located on the chromosome from getting deleted as cells continue to divide. In humans, a six base pair sequence, TTAGGG, is repeated 100 to 1000 times. The discovery of the enzyme telomerase (Figure 1) helped in the understanding of how chromosome ends are maintained. The telomerase enzyme contains a catalytic part and a built-in RNA template. It attaches to the end of the chromosome, and complementary bases to the RNA template are added on the 3′ end of the DNA strand. Once the 3′ end of the lagging strand template is sufficiently elongated, DNA polymerase can add the nucleotides complementary to the ends of the chromosomes. Thus, the ends of the chromosomes are replicated.
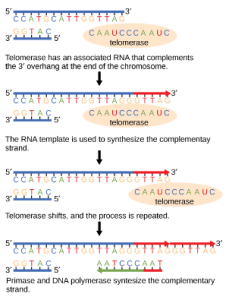
Telomerase is typically active in germ cells and adult stem cells. It is not active in adult somatic cells. For her discovery of telomerase and its action, Elizabeth Blackburn (Figure 2) received the Nobel Prize for Medicine and Physiology in 2009.

Telomerase and Aging
Cells that undergo cell division continue to have their telomeres shortened because most somatic cells do not make telomerase. This essentially means that telomere shortening is associated with aging. With the advent of modern medicine, preventative health care, and healthier lifestyles, the human life span has increased, and there is an increasing demand for people to look younger and have a better quality of life as they grow older.
In 2010, scientists found that telomerase can reverse some age-related conditions in mice. This may have potential in regenerative medicine (Jaskelioff, 2011). Telomerase-deficient mice were used in these studies; these mice have tissue atrophy, stem cell depletion, organ system failure, and impaired tissue injury responses. Telomerase reactivation in these mice caused extension of telomeres, reduced DNA damage, reversed neurodegeneration, and improved the function of the testes, spleen, and intestines. Thus, telomere reactivation may have potential for treating age-related diseases in humans.
Cancer is characterized by uncontrolled cell division of abnormal cells. The cells accumulate mutations, proliferate uncontrollably, and can migrate to different parts of the body through a process called metastasis. Scientists have observed that cancerous cells have considerably shortened telomeres and that telomerase is active in these cells. Interestingly, only after the telomeres were shortened in the cancer cells did the telomerase become active. If the action of telomerase in these cells can be inhibited by drugs during cancer therapy, then the cancerous cells could potentially be stopped from further division.
References
Unless otherwise noted, images on this page are licensed under CC-BY 4.0 by OpenStax.
OpenStax, Concepts of Biology. OpenStax CNX. May 18, 2016 http://cnx.org/contents/s8Hh0oOc@9.10:2ousESf0@5/DNA-Replication
Jaskelioff et al., 2011 Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 469: 102-7.

